








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (11): 227-237.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0280
Previous Articles Next Articles
GUO Yu-fei( ), YAN Rong-mei, ZHANG Xiao-ru, CAO Wei, LIU Hao(
), YAN Rong-mei, ZHANG Xiao-ru, CAO Wei, LIU Hao( )
)
Received:2022-03-06
Online:2022-11-26
Published:2022-12-01
Contact:
LIU Hao
E-mail:guoyufeis@163.com;liuhao@tust.edu.cn
GUO Yu-fei, YAN Rong-mei, ZHANG Xiao-ru, CAO Wei, LIU Hao. Metabolic Engineering Modification of Aspergillus niger for the Production of D-glucaric Acid[J]. Biotechnology Bulletin, 2022, 38(11): 227-237.
| 年份Year | 菌株Strain | 基因改造Genes madified | 碳源Carbon source | 产量Yield/(g·L-1) | 引用文献Reference |
|---|---|---|---|---|---|
| 2009 | E. coli BL21 Star(DE3) | ino1,MIOX,udh | Glucose | 1.13 | [ |
| 2010 | E. coli BL21 Star(DE3) | ino1,MIOX,udh | Glucose | 2.50 | [ |
| 2014 | E. coli MG1655 | ino1,SUMO-MIOX,udh | Myo-inositol | 4.85 | [ |
| 2016 | S. cerevisiae CEN. PK2-1D | ino1,inm,MIOX,udh | Glucose | 1.60 | [ |
| 2017 | E. coli L19S | ino1,MIOX,udh | Glucose | 0.80 | [ |
| 2017 | S. cerevisiae CEN. PK2-1C | MIOX,udh,ino1,pfk1 | Glucose | 0.23 | [ |
| 2018 | E. coli BL21 Star(DE3) | cscB,cscA,cscK,ino1, MIOX,udh,suhB | Sucrose | 1.42 | [ |
| 2018 | Pichia pastoris GS115 | MIOX,udh | Glucose | 2.60 | [ |
| 2018 | S. cerevisiae BY4741 | ino1,MIOX,udh | Glucose,myo-inositol | 6.00 | [ |
| 2020 | E. coli BL21 Star(DE3) | ino1,MIOX,udh,suhB | Glucose | 5.35 | [ |
| 2020 | S. cerevisiae | ino1,inm,MIOX,udh,vgb | Glucose | 6.38 | [ |
Table 1 Research progress of producing gluconic acid by microbial fermentation
| 年份Year | 菌株Strain | 基因改造Genes madified | 碳源Carbon source | 产量Yield/(g·L-1) | 引用文献Reference |
|---|---|---|---|---|---|
| 2009 | E. coli BL21 Star(DE3) | ino1,MIOX,udh | Glucose | 1.13 | [ |
| 2010 | E. coli BL21 Star(DE3) | ino1,MIOX,udh | Glucose | 2.50 | [ |
| 2014 | E. coli MG1655 | ino1,SUMO-MIOX,udh | Myo-inositol | 4.85 | [ |
| 2016 | S. cerevisiae CEN. PK2-1D | ino1,inm,MIOX,udh | Glucose | 1.60 | [ |
| 2017 | E. coli L19S | ino1,MIOX,udh | Glucose | 0.80 | [ |
| 2017 | S. cerevisiae CEN. PK2-1C | MIOX,udh,ino1,pfk1 | Glucose | 0.23 | [ |
| 2018 | E. coli BL21 Star(DE3) | cscB,cscA,cscK,ino1, MIOX,udh,suhB | Sucrose | 1.42 | [ |
| 2018 | Pichia pastoris GS115 | MIOX,udh | Glucose | 2.60 | [ |
| 2018 | S. cerevisiae BY4741 | ino1,MIOX,udh | Glucose,myo-inositol | 6.00 | [ |
| 2020 | E. coli BL21 Star(DE3) | ino1,MIOX,udh,suhB | Glucose | 5.35 | [ |
| 2020 | S. cerevisiae | ino1,inm,MIOX,udh,vgb | Glucose | 6.38 | [ |
| 菌株Strain | 特征描述Characteristics | 来源Source |
|---|---|---|
| Escherichia coli JM109 | - | 本实验室保存 |
| Agrobacterium tumerfaciens AGL-1 | - | 本实验室保存 |
| S834 | A.niger ATCC1015,Tet-on::cre,ΔoahA,ΔcexA | 本实验室保存 |
| S985 | ΔoahA,ΔcexA,OEppudh | 本研究 |
| S1223 | ΔoahA,ΔcexA,OEppudh,OEanmioxA | 本研究 |
| S1877 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1 | 本研究 |
| S2086 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1,OEaninoA | 本研究 |
| S2390 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1,OEaninoA,OEllnox | 本研究 |
| S2603 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1,OEaninoA,OEllnox,RNAipfkA | 本研究 |
| S2635 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1,OEaninoA,OEllnox,RNAipfkA,RNAizwf | 本研究 |
Table 2 Strains used in this study
| 菌株Strain | 特征描述Characteristics | 来源Source |
|---|---|---|
| Escherichia coli JM109 | - | 本实验室保存 |
| Agrobacterium tumerfaciens AGL-1 | - | 本实验室保存 |
| S834 | A.niger ATCC1015,Tet-on::cre,ΔoahA,ΔcexA | 本实验室保存 |
| S985 | ΔoahA,ΔcexA,OEppudh | 本研究 |
| S1223 | ΔoahA,ΔcexA,OEppudh,OEanmioxA | 本研究 |
| S1877 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1 | 本研究 |
| S2086 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1,OEaninoA | 本研究 |
| S2390 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1,OEaninoA,OEllnox | 本研究 |
| S2603 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1,OEaninoA,OEllnox,RNAipfkA | 本研究 |
| S2635 | ΔoahA,ΔcexA,OEppudh,OEanmioxA,OEscJEN1,OEaninoA,OEllnox,RNAipfkA,RNAizwf | 本研究 |
| 质粒Plasmid | 特征描述Characteristics | 来源Source |
|---|---|---|
| pLH454 | loxP-hph-loxP,gpdA promoter,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH509 | loxP-hph-loxP,pkiA promoter,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH756 | loxP-hph-loxP,mbfA promoter,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH1418 | loxP-hph-loxP,glaA promoter,gfp loop,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH1453 | loxP-hph-loxP,pkiA promoter,gfp loop,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH690 | loxP-hph-loxP,gpdA promoter,ppudh gene,trpC terminator,hygr,kanr | 本研究 |
| pLH737 | loxP-hph-loxP,pkiA promoter,anmioxA gene,trpC terminator,hygr,kanr | 本研究 |
| pLH782 | loxP-hph-loxP,gpdA promoter,scJEN1 gene,trpC terminator,hygr,kanr | 本研究 |
| pLH1020 | loxP-hph-loxP,gpdA promoter,aninoA gene,trpC terminator,hygr,kanr | 本研究 |
| pLH1317 | loxP-hph-loxP,mbfA promoter,llnox gene,trpC terminator,hygr,kanr | 本研究 |
| pLH1421 | loxP-hph-loxP,glaA promoter,sense sequence of zwf gene,gfp loop,antisense sequence of zwf gene,trpC terminator,hygr,kanr | 本研究 |
| pLH1454 | loxP-hph-loxP,pkiA promoter,sense sequence of pfkA gene,gfp loop,antisense sequence of pfkA gene,trpC terminator,hygr,kanr | 本研究 |
Table 3 Plasmids used in this study
| 质粒Plasmid | 特征描述Characteristics | 来源Source |
|---|---|---|
| pLH454 | loxP-hph-loxP,gpdA promoter,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH509 | loxP-hph-loxP,pkiA promoter,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH756 | loxP-hph-loxP,mbfA promoter,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH1418 | loxP-hph-loxP,glaA promoter,gfp loop,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH1453 | loxP-hph-loxP,pkiA promoter,gfp loop,trpC terminator,hygr,kanr | 本实验室保存 |
| pLH690 | loxP-hph-loxP,gpdA promoter,ppudh gene,trpC terminator,hygr,kanr | 本研究 |
| pLH737 | loxP-hph-loxP,pkiA promoter,anmioxA gene,trpC terminator,hygr,kanr | 本研究 |
| pLH782 | loxP-hph-loxP,gpdA promoter,scJEN1 gene,trpC terminator,hygr,kanr | 本研究 |
| pLH1020 | loxP-hph-loxP,gpdA promoter,aninoA gene,trpC terminator,hygr,kanr | 本研究 |
| pLH1317 | loxP-hph-loxP,mbfA promoter,llnox gene,trpC terminator,hygr,kanr | 本研究 |
| pLH1421 | loxP-hph-loxP,glaA promoter,sense sequence of zwf gene,gfp loop,antisense sequence of zwf gene,trpC terminator,hygr,kanr | 本研究 |
| pLH1454 | loxP-hph-loxP,pkiA promoter,sense sequence of pfkA gene,gfp loop,antisense sequence of pfkA gene,trpC terminator,hygr,kanr | 本研究 |
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| Primers related to ppudh gene | |
| YZ-ppudh-F | ACCACCACCCCCTTCAAT |
| YZ-ppudh-R | TCAGTGATGGTGGTGATGGT |
| qPCR-ppudh-F | GCAACCACACCATCGGCTTCTAC |
| qPCR-ppudh-R | CCATAGCGGTCGAAGTAGAAGGAGG |
| Primers related to anmioxA gene | |
| anmioxA-F | ATCAATCATCCGTCAAGATGGAATTCGCACCAGTCGCTGTGTCTCCT |
| anmioxA-R | CCAGATCTCTGCAGGGTACCGAGCTCCTACCACTTGATCACCTTGTTGG |
| qPCR-anmioxA-F | GAAGCTCAACACCCTCATCGACG |
| qPCR-anmioxA-R | ATGAGACCGGTCAGCTGCATC |
| Primers related to scJEN1 gene | |
| YZ-scJEN1-F | ATGTCCTCCAGCATTACCGAT |
| YZ-scJEN1-R | GACCTTGGCGTAATCTTCCTT |
| qPCR-scJEN1-F | GACCACGAGAAGCTGTACCATAACCC |
| qPCR-scJEN1-R | GGGCTTATCTTCTTCGTCCTCCTCG |
| Primers related to aninoA gene | |
| aninoA-F | CACATCTAAACAATGGAATTCATGGCTCCCCACGCAAGC |
| aninoA-R | AGTGGATCCCTGCAGGGTACCCTAGAACAGCTTGTGCTCCAG |
| qPCR-aninoA-F | GGCCACCAACTACCACTTCAAGG |
| qPCR-aninoA-R | CCACAGAGCCGTAGTAGTTGGAGG |
| Primers related to llnox gene | |
| YZ-llnox-F | TCATCGGCACCAACCACG |
| YZ-llnox-R | ACGGTCATGTAGTTGTAGGGG |
| qPCR-llnox-F | ATCTGGTCATCAACTGCATCGG |
| qPCR-llnox-R | CCAGGGCGATGTAGGTGAAATC |
| Primers related to zwf gene | |
| zwf-S-F | TACACCTCAGCAATGGAGCTCAGAAGGAGCAGAACAGAATCT |
| zwf-S-R | GCTCCTGGAAGATCTGAGCTCATCCAGAGACTTTCCGTACT |
| zwf-A-F | CGCATCGAGCTGAAGGGTACCATCCAGAGACTTTCCGTACT |
| zwf-A-R | AAGTGGATCGCATGCGGTACCAGAAGGAGCAGAACAGAATCT |
| YZ-PglaA-F | ACCTGCGTTATAGCTTCCCG |
| YZ-gfp-F | AGGAGCGCACCATCTTCTTC |
| YZ-gfp-R | CTCGATGCGGTTCACCAGG |
| YZ-TtrpC-R | CTCCGGAGCTGACATCGAC |
| qPCR-zwf-F | GAACGAGAGGTGGGACGGTG |
| qPCR-zwf-R | CAGGCAGCTTGGAGTTCATCTTG |
| Primers related to pfkA gene | |
| pfkA-S-F | ATCAATCATCCGTCAAGATGGAATTCACTATGATGACACCCCGATCT |
| pfkA-S-R | GGTGCGCTCCTGGAGGTACCGAGCTCCAAAGTTGTCACGCAGGAAGT |
| pfkA-A-F | TGAACCGCATCGAGCTGAAGCTGCAGCAAAGTTGTCACGCAGGAAGT |
| pfkA-A-R | TGATTTCAGTAACGTTAAGTCTGCAGACTATGATGACACCCCGATCT |
| YZ-PpkiA-F | GCGAGGAGAAAATTCAGCACC |
| YZ-gfp-F | AGGAGCGCACCATCTTCTTC |
| YZ-gfp-R | CTCGATGCGGTTCACCAGG |
| YZ-TtrpC-R | CTCCGGAGCTGACATCGAC |
| qPCR-pfkA-F | ACCATTTCCAACAACGTGCCG |
| qPCR-pfkA-R | TGTCACGCAGGAAGTCAATGTCG |
| Primers related to actA gene | |
| qPCR-actA-F | TCCTCACCCTCAGATACCC |
| qPCR-actA-R | CACCGTCACCAGAGTCCA |
Table 4 Primers used in this study
| 引物Primer | 序列Sequence(5'-3') |
|---|---|
| Primers related to ppudh gene | |
| YZ-ppudh-F | ACCACCACCCCCTTCAAT |
| YZ-ppudh-R | TCAGTGATGGTGGTGATGGT |
| qPCR-ppudh-F | GCAACCACACCATCGGCTTCTAC |
| qPCR-ppudh-R | CCATAGCGGTCGAAGTAGAAGGAGG |
| Primers related to anmioxA gene | |
| anmioxA-F | ATCAATCATCCGTCAAGATGGAATTCGCACCAGTCGCTGTGTCTCCT |
| anmioxA-R | CCAGATCTCTGCAGGGTACCGAGCTCCTACCACTTGATCACCTTGTTGG |
| qPCR-anmioxA-F | GAAGCTCAACACCCTCATCGACG |
| qPCR-anmioxA-R | ATGAGACCGGTCAGCTGCATC |
| Primers related to scJEN1 gene | |
| YZ-scJEN1-F | ATGTCCTCCAGCATTACCGAT |
| YZ-scJEN1-R | GACCTTGGCGTAATCTTCCTT |
| qPCR-scJEN1-F | GACCACGAGAAGCTGTACCATAACCC |
| qPCR-scJEN1-R | GGGCTTATCTTCTTCGTCCTCCTCG |
| Primers related to aninoA gene | |
| aninoA-F | CACATCTAAACAATGGAATTCATGGCTCCCCACGCAAGC |
| aninoA-R | AGTGGATCCCTGCAGGGTACCCTAGAACAGCTTGTGCTCCAG |
| qPCR-aninoA-F | GGCCACCAACTACCACTTCAAGG |
| qPCR-aninoA-R | CCACAGAGCCGTAGTAGTTGGAGG |
| Primers related to llnox gene | |
| YZ-llnox-F | TCATCGGCACCAACCACG |
| YZ-llnox-R | ACGGTCATGTAGTTGTAGGGG |
| qPCR-llnox-F | ATCTGGTCATCAACTGCATCGG |
| qPCR-llnox-R | CCAGGGCGATGTAGGTGAAATC |
| Primers related to zwf gene | |
| zwf-S-F | TACACCTCAGCAATGGAGCTCAGAAGGAGCAGAACAGAATCT |
| zwf-S-R | GCTCCTGGAAGATCTGAGCTCATCCAGAGACTTTCCGTACT |
| zwf-A-F | CGCATCGAGCTGAAGGGTACCATCCAGAGACTTTCCGTACT |
| zwf-A-R | AAGTGGATCGCATGCGGTACCAGAAGGAGCAGAACAGAATCT |
| YZ-PglaA-F | ACCTGCGTTATAGCTTCCCG |
| YZ-gfp-F | AGGAGCGCACCATCTTCTTC |
| YZ-gfp-R | CTCGATGCGGTTCACCAGG |
| YZ-TtrpC-R | CTCCGGAGCTGACATCGAC |
| qPCR-zwf-F | GAACGAGAGGTGGGACGGTG |
| qPCR-zwf-R | CAGGCAGCTTGGAGTTCATCTTG |
| Primers related to pfkA gene | |
| pfkA-S-F | ATCAATCATCCGTCAAGATGGAATTCACTATGATGACACCCCGATCT |
| pfkA-S-R | GGTGCGCTCCTGGAGGTACCGAGCTCCAAAGTTGTCACGCAGGAAGT |
| pfkA-A-F | TGAACCGCATCGAGCTGAAGCTGCAGCAAAGTTGTCACGCAGGAAGT |
| pfkA-A-R | TGATTTCAGTAACGTTAAGTCTGCAGACTATGATGACACCCCGATCT |
| YZ-PpkiA-F | GCGAGGAGAAAATTCAGCACC |
| YZ-gfp-F | AGGAGCGCACCATCTTCTTC |
| YZ-gfp-R | CTCGATGCGGTTCACCAGG |
| YZ-TtrpC-R | CTCCGGAGCTGACATCGAC |
| qPCR-pfkA-F | ACCATTTCCAACAACGTGCCG |
| qPCR-pfkA-R | TGTCACGCAGGAAGTCAATGTCG |
| Primers related to actA gene | |
| qPCR-actA-F | TCCTCACCCTCAGATACCC |
| qPCR-actA-R | CACCGTCACCAGAGTCCA |
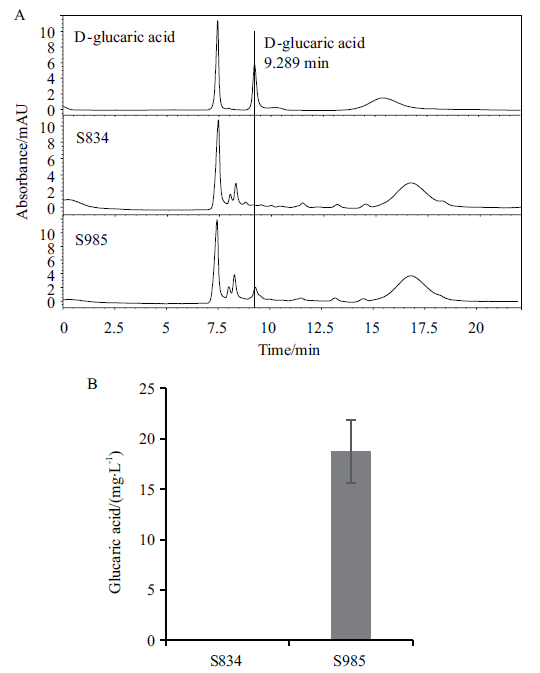
Fig. 2 Effect of ppudh gene expression on glucaric acid production A:HPLC chromatogram of 100 mg/L glucaric acid standard and different strains fermentation. B:Glucaric acid yield of the modified strain
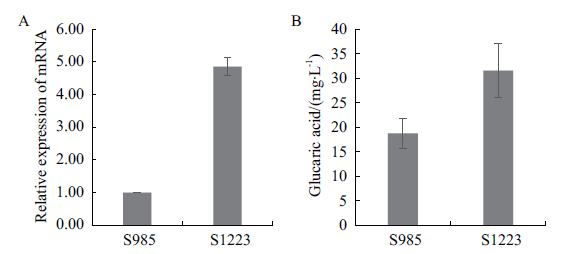
Fig. 3 Effect of anmioxA gene expression on glucaric acid production A:The relative expression of anmioxA gene in the modified strain. B:Glucaric acid yield of the modified strain
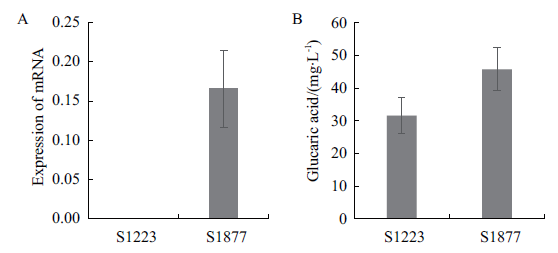
Fig. 4 Effect of scJEN1 gene expression on glucaric acid production A:Expression of scJEN1 gene in the modified strain. B:Glucaric acid yield of the modified strain
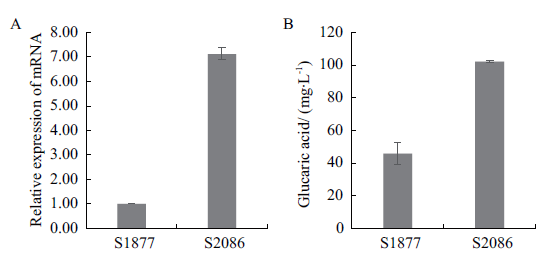
Fig. 5 Effect of aninoA gene expression on glucaric acid production A:Relative expression of aninoA gene in the modified strain. B:Glucaric acid yield of the modified strain
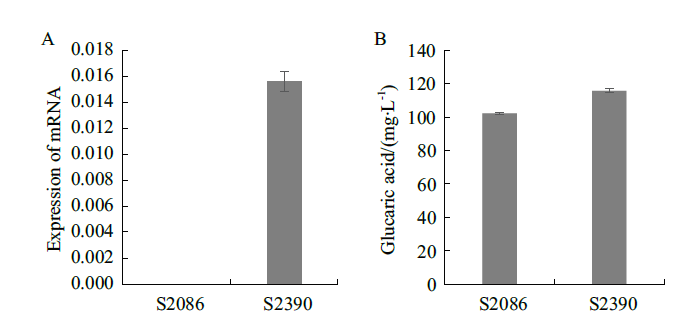
Fig. 6 Effects of llnox gene expression on glucaric acid production A:Expression of llnox gene in the modified strain;B:Glucaric acid yield of the modified strain
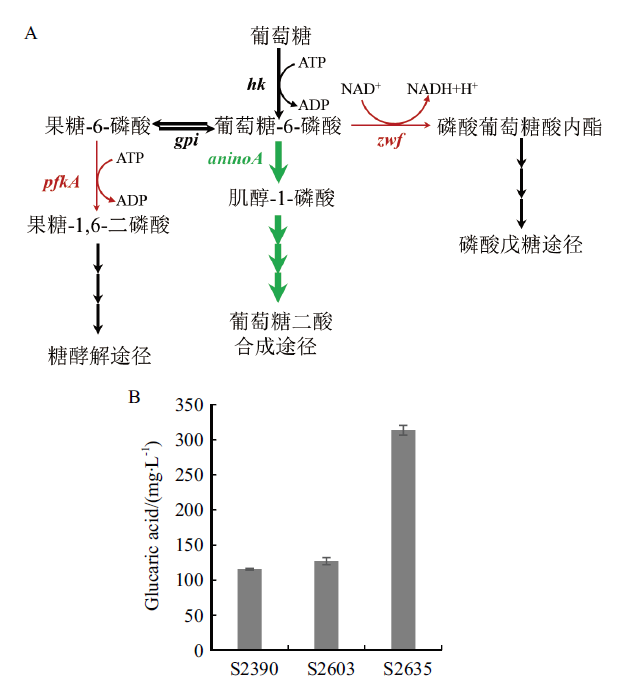
Fig. 7 Effect of interfering pfkA and zwf gene on glucaric acid production A:Branch metabolism of glucose-6-phosphate(Red arrows indicate weakened expression and green bold arrows indicate enhanced expression). B:Glucaric acid yield of the modified strain
| [1] |
Walaszek Z, Szemraj J, Hanausek M, et al. D-Glucaric acid content of various fruits and vegetables and cholesterol-lowering effects of dietary d-glucarate in the rat[J]. Nutr Res, 1996, 16(4):673-681.
doi: 10.1016/0271-5317(96)00045-0 URL |
| [2] | Werpy T, Petersen G. Top Value Added Chemicals from Biomass:Volume I -- Results of Screening for Potential Candidates from Sugars and Synthesis Gas[R]. Office of Scientific and Technical Information(OSTI), 2004. |
| [3] |
Marsh CA. Metabolism of d-glucuronolactone in mammalian systems. 2. Conversion of d-glucuronolactone into d-glucaric acid by tissue preparations[J]. Biochem J, 1963, 87(1):82-90.
pmid: 16748999 |
| [4] | Zółtaszek R, Hanausek M, Kiliańska ZM, et al. The biological role of D-glucaric acid and its derivatives:potential use in medicine[J]. Postepy Hig Med Dosw(Online), 2008, 62:451-462. |
| [5] |
Walaszek Z. Potential use of D-glucaric acid derivatives in cancer prevention[J]. Cancer Lett, 1990, 54(1/2):1-8.
doi: 10.1016/0304-3835(90)90083-A URL |
| [6] | 余作龙, 苗江月, 曹飞, 等. 葡萄糖二酸的生物炼制及应用研究进展[J]. 化工进展, 2011, 30(11):2502-2508, 2535. |
| Yu ZL, Miao JY, Cao F, et al. Research progress in preparation and application of bio-refined D-glucaric acid[J]. Chem Ind Eng Prog, 2011, 30(11):2502-2508, 2535. | |
| [7] | 仇钰莹, 方芳, 堵国成, 等. 葡萄糖二酸研究进展[J]. 生物工程学报, 2015, 31(4):481-490. |
| Qiu YY, Fang F, Du GC, et al. Progress in glucaric acid[J]. Chin J Biotechnol, 2015, 31(4):481-490. | |
| [8] | Kiely DE, Hash KR, Sr. Method of oxidation using nitric acid:US9162959[P]. 2015-10-20. |
| [9] |
Ibert M, Fuertès P, Merbouh N, et al. Improved preparative electrochemical oxidation of d-glucose to d-glucaric acid[J]. Electrochimica Acta, 2010, 55(10):3589-3594.
doi: 10.1016/j.electacta.2009.11.041 URL |
| [10] | 朱俏俏, 庄军平, 余开荣. 生物质基葡萄糖二酸的研究与展望[J]. 工业催化, 2019, 27(9):13-18. |
| Zhu QQ, Zhuang JP, Yu KR. Current status and future prospect of biomass-based glucaric acid[J]. Ind Catal, 2019, 27(9):13-18. | |
| [11] | Pamuk V, Yılmaz M, Alıcılar A. The preparation of D-glucaric acid by oxidation of molasses in packed beds[J]. J Chem Technol Biotechnol, 2001, 76(2):186-190. |
| [12] |
Moon TS, Yoon SH, Lanza AM, et al. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli[J]. Appl Environ Microbiol, 2009, 75(3):589-595.
doi: 10.1128/AEM.00973-08 URL |
| [13] |
Moon TS, Dueber JE, Shiue E, et al. Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli[J]. Metab Eng, 2010, 12(3):298-305.
doi: 10.1016/j.ymben.2010.01.003 URL |
| [14] |
Gupta A, Hicks MA, Manchester SP, et al. Porting the synthetic D-glucaric acid pathway from Escherichia coli to Saccharomyces cerevisiae[J]. Biotechnol J, 2016, 11(9):1201-1208.
doi: 10.1002/biot.201500563 URL |
| [15] | 巩旭, 刘叶, 王毳, 等. 代谢工程改造酿酒酵母合成葡萄糖二酸[J]. 生物工程学报, 2017, 33(2):228-236. |
| Gong X, Liu Y, Wang C, et al. Metabolic engineering of Saccharomyces cerevisiae for production of glucaric acid[J]. Chin J Biotechnol, 2017, 33(2):228-236. | |
| [16] | 刘叶, 巩旭, 康振, 等. 代谢工程改造毕赤酵母生产葡萄糖二酸[J]. 食品与生物技术学报, 2018, 37(9):955-961. |
| Liu Y, Gong X, Kang Z, et al. Metabolic engineering of Pichia pastoris for production of glucaric acid[J]. J Food Sci Biotechnol, 2018, 37(9):955-961. | |
| [17] |
Shiue E, Prather KLJ. Improving D-glucaric acid production from myo-inositol in E. coli by increasing MIOX stability and myo-inositol transport[J]. Metab Eng, 2014, 22:22-31.
doi: 10.1016/j.ymben.2013.12.002 pmid: 24333274 |
| [18] |
Gupta A, Reizman IM, Reisch CR, et al. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit[J]. Nat Biotechnol, 2017, 35(3):273-279.
doi: 10.1038/nbt.3796 pmid: 28191902 |
| [19] |
Qu YN, Yan HJ, Guo Q, et al. Biosynthesis of D-glucaric acid from sucrose with routed carbon distribution in metabolically engineered Escherichia coli[J]. Metab Eng, 2018, 47:393-400.
doi: S1096-7176(17)30484-6 pmid: 29715517 |
| [20] |
Chen N, Wang JY, Zhao YY, et al. Metabolic engineering of Saccharomyces cerevisiae for efficient production of glucaric acid at high titer[J]. Microb Cell Fact, 2018, 17(1):67.
doi: 10.1186/s12934-018-0914-y pmid: 29729665 |
| [21] |
Su HH, Peng F, Ou XY, et al. Combinatorial synthetic pathway fine-tuning and cofactor regeneration for metabolic engineering of Escherichia coli significantly improve production of D-glucaric acid[J]. N Biotechnol, 2020, 59:51-58.
doi: 10.1016/j.nbt.2020.03.004 URL |
| [22] |
Zhang X, Xu C, Liu YL, et al. Enhancement of glucaric acid production in Saccharomyces cerevisiae by expressing Vitreoscilla hemoglobin[J]. Biotechnol Lett, 2020, 42(11):2169-2178.
doi: 10.1007/s10529-020-02966-2 pmid: 32691185 |
| [23] |
Schuster E, Dunn-Coleman N, Frisvad JC, et al. On the safety of Aspergillus niger-a review[J]. Appl Microbiol Biotechnol, 2002, 59(4/5):426-435.
doi: 10.1007/s00253-002-1032-6 URL |
| [24] |
Xu YX, Shan L, Zhou YT, et al. Development of a Cre-loxP-based genetic system in Aspergillus niger ATCC1015 and its application to construction of efficient organic acid-producing cell factories[J]. Appl Microbiol Biotechnol, 2019, 103(19):8105-8114.
doi: 10.1007/s00253-019-10054-3 URL |
| [25] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [26] | 秦俊文, 谢琪璇, 蔡冬青, 等. shRNA慢病毒质粒的构建技巧[J]. 中国生物工程杂志, 2011, 31(3):124-127. |
| Qin JW, Xie QX, Cai DQ, et al. Construction techniques of shRNA lentivirus plasmid[J]. China Biotechnol, 2011, 31(3):124-127. | |
| [27] |
Chang SS, Zhang ZY, Liu Y. RNA interference pathways in fungi:mechanisms and functions[J]. Annu Rev Microbiol, 2012, 66:305-323.
doi: 10.1146/annurev-micro-092611-150138 URL |
| [28] |
Cairns TC, Nai C, Meyer V. How a fungus shapes biotechnology:100 years of Aspergillus niger research[J]. Fungal Biol Biotechnol, 2018, 5:13.
doi: 10.1186/s40694-018-0054-5 URL |
| [29] |
Andersen MR, Lehmann L, Nielsen J. Systemic analysis of the response of Aspergillus niger to ambient pH[J]. Genome Biol, 2009, 10(5):R47.
doi: 10.1186/gb-2009-10-5-r47 URL |
| [30] | 石慧, 陈卓逐, 阚建全. 大肠杆菌在食品加工贮藏中胁迫响应机制的研究进展[J]. 食品科学, 2016, 37(9):250-257. |
|
Shi H, Chen ZZ, Kan JQ. Progress in research on stress response in Escherichia coli during food processing and storage[J]. Food Sci, 2016, 37(9):250-257.
doi: 10.1111/j.1365-2621.1972.tb05828.x URL |
|
| [31] |
Yoon SH, Moon TS, Iranpour P, et al. Cloning and characterization of uronate dehydrogenases from two pseudomonads and Agrobacterium tumefaciens strain C58[J]. J Bacteriol, 2009, 191(5):1565-1573.
doi: 10.1128/JB.00586-08 URL |
| [32] | Ribas D, Sá-Pessoa J, Soares-Silva I, et al. Yeast as a tool to express sugar acid transporters with biotechnological interest[J]. FEMS Yeast Res, 2017, 17(2):2017 Mar 1;17(2). |
| [33] |
Shi XC, Zou YN, Chen Y, et al. A water-forming NADH oxidase regulates metabolism in anaerobic fermentation[J]. Biotechnol Biofuels, 2016, 9:103.
doi: 10.1186/s13068-016-0517-y URL |
| [1] | XUE Xian-li, WANG Jing-ran, BI Hang-hang, WANG De-pei. Effect of Spt7 Overexpression of on the Growth and Stress Resistance of Aspergillus niger [J]. Biotechnology Bulletin, 2022, 38(5): 112-122. |
| [2] | LIU Xiao-mei, WANG Dong-xin, ZHANG Chun, WEI Shuang-shi. Inhibition of AAV-mediated RNAi to SARS-CoV-2 S Gene Expression [J]. Biotechnology Bulletin, 2022, 38(3): 188-193. |
| [3] | PAN Yin-lai, QIU Chun-hui, WANG Yi-lei, ZHANG Zi-ping. Development of RNA Drugs and Its Application in Aquaculture [J]. Biotechnology Bulletin, 2021, 37(2): 203-215. |
| [4] | MENG Xiao-jian, YU Jian-dong, ZHENG Xiao-mei, ZHENG Ping, LI Zhi-min, SUN Ji-bin, YE Qin. Regulations of Small-molecules Metabolites on Hexokinase and Pyruvate Kinase in Aspergillus niger [J]. Biotechnology Bulletin, 2021, 37(12): 180-190. |
| [5] | DENG Pu-rong, LIU Yong-bo. Review on the Synergistic Insect-resistant Application of RNAi and Bt-transgenic Technologies [J]. Biotechnology Bulletin, 2021, 37(10): 216-224. |
| [6] | XU Xue-liang, WANG Fen-shan, LIU Zi-rong, FAN Lin-juan, JI Xiang-yun, JIANG Jie-xian, YAO Ying-juan. Research Progress of RNA Interference Technology in the Field of Entomology [J]. Biotechnology Bulletin, 2021, 37(1): 255-261. |
| [7] | SU Jie, GUO Rong-qi, GAO Yang, YU Xiu-min, LI Guo-jing, WANG Rui-gang. Response to NaCl and ABA in Arabidopsis thaliana of the Double Silent Gene VHA-c2&c4 [J]. Biotechnology Bulletin, 2020, 36(7): 48-54. |
| [8] | SONG Hua-li, SUN Xiao-ying, KONG Xiang-hui, LI Li, PEI Chao. Application of RNA Interference Technology in Antiviral and Antiparasitic Research of Aquatic Animals [J]. Biotechnology Bulletin, 2020, 36(2): 193-205. |
| [9] | HAN Cui-cui, LIU Li-kun, WANG Yu-chun, YANG Ying, LIU Ji-cheng, ZHOU Zhong-guang. Construction of TOX3 Gene Lentiviral RNA Interference Vector and Effect on Proliferation of Human Breast Cancer Cells ZR-75-1 [J]. Biotechnology Bulletin, 2019, 35(7): 141-147. |
| [10] | WANG Jia-yue, LIU Xiang-nan, PENG Kang-li, ZHAO Bo. Construction and Identification of Lentiviral Vector for RNA Interference of USE1 Gene [J]. Biotechnology Bulletin, 2019, 35(3): 117-122. |
| [11] | Su Zijing, Li Qiaoling, Huang Cheng, Xie Chengjian, Yang Xingyong. RNAi Technology and Its Application in Fungal Gene Functional Studies [J]. Biotechnology Bulletin, 2015, 31(8): 50-58. |
| [12] | Wen Xianchun, Han Cuicui, Zhao Yuesheng, Yu Haitao, Li Chengchong, Yue Liling. Construction of FUT8 Gene Lentiviral RNA Interference Vector and Regulation on Proliferation of Human Breast Cancer Cells MCF-7 [J]. Biotechnology Bulletin, 2015, 31(5): 231-236. |
| [13] | Qiu Xier, Zhu Dongfa, Zhou Yanqi, Liu Zhiye, Xie Xi. Progress of Research and Application of the RNA Interference Technology in Crustacean [J]. Biotechnology Bulletin, 2015, 31(3): 57-63. |
| [14] | Deng Shuai, Zhang Tingting, Wang Ruru, Liu Yu, Zhang Yuanhu. Advances on the Research of Non-cell-autonomous Small RNAs in Plants [J]. Biotechnology Bulletin, 2015, 31(10): 16-23. |
| [15] | Shi Meng, Liu Xiaoning, Ma Ji. RNA Interference of Antifreeze Protein Gene in Tenebrio molitor Mediated by Bacterially Expressed dsRNA [J]. Biotechnology Bulletin, 2014, 0(8): 113-119. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||