








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (11): 350-359.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0631
Previous Articles Next Articles
WU Bai-zeng1,2( ), HE Qi2,3, YAO Fang-jie1,3(
), HE Qi2,3, YAO Fang-jie1,3( ), ZHAO Meng-ran2(
), ZHAO Meng-ran2( )
)
Received:2023-07-03
Online:2023-11-26
Published:2023-12-20
Contact:
YAO Fang-jie, ZHAO Meng-ran
E-mail:1772696751@qq.com;yaofj@aliyun.com;zhaomengran@caas.cn
WU Bai-zeng, HE Qi, YAO Fang-jie, ZHAO Meng-ran. Identification of Lactate Dehydrogenase in Pleurotus ostreatus and Heat Stress Expression Analysis of Mycelium[J]. Biotechnology Bulletin, 2023, 39(11): 350-359.
| 引物Primer | 正向序列Forward sequence(5'-3') | 反向序列Reverse sequence(5'-3') |
|---|---|---|
| qPCR-ldh1 | GGTCCTGCTTTGTCTCGTAG | GGGAAAGGATGGAGTACAGATG |
| qPCR-ldh2 | AGAGGAATTGGAAAGACATGGG | ATACGGAATGACGGGCATAAG |
| qPCR-ldh3 | CTTGGGAGATGTGATACTGGG | GTTCTCGGTTTGAATTCGCTG |
| qPCR-ldh4 | GCGATTAGTGGCAAGGTTTAC | CATCCAACATGAAATCGGCG |
| qPCR-ldh5 | CACCCGCTCTACCAATTCTG | GCCATTCCTTTACGCTCAATC |
| qPCR-ldh6 | TGCTTCTACCCGTTTCTGTC | ATATGACCCAATACATCCACCG |
| qPCR-ldh7 | TGAACTTGCGTCTACTTGGTG | CAGGCGGAGAGGTTGATATG |
| qPCR-ldh8 | GGCGGAGGTGTCAAGATTC | ATGGCGGAACGAAGGATATG |
Table 1 Primers for qPCR
| 引物Primer | 正向序列Forward sequence(5'-3') | 反向序列Reverse sequence(5'-3') |
|---|---|---|
| qPCR-ldh1 | GGTCCTGCTTTGTCTCGTAG | GGGAAAGGATGGAGTACAGATG |
| qPCR-ldh2 | AGAGGAATTGGAAAGACATGGG | ATACGGAATGACGGGCATAAG |
| qPCR-ldh3 | CTTGGGAGATGTGATACTGGG | GTTCTCGGTTTGAATTCGCTG |
| qPCR-ldh4 | GCGATTAGTGGCAAGGTTTAC | CATCCAACATGAAATCGGCG |
| qPCR-ldh5 | CACCCGCTCTACCAATTCTG | GCCATTCCTTTACGCTCAATC |
| qPCR-ldh6 | TGCTTCTACCCGTTTCTGTC | ATATGACCCAATACATCCACCG |
| qPCR-ldh7 | TGAACTTGCGTCTACTTGGTG | CAGGCGGAGAGGTTGATATG |
| qPCR-ldh8 | GGCGGAGGTGTCAAGATTC | ATGGCGGAACGAAGGATATG |
| 名称 Name | 互补DNA/基因组DNA cDNA/gDNA/bp | 分子量 Molecular weight/kD | 等电点 pI |
|---|---|---|---|
| D-ldh1 | 1473/2114 | 52.91 | 4.88 |
| D-ldh2 | 1692/2459 | 61.34 | 4.84 |
| D-ldh3 | 1776/2603 | 63.92 | 4.82 |
| L-ldh4 | 711/820 | 25.18 | 5.05 |
| L-ldh5 | 1485/2158 | 54.54 | 4.86 |
| L-ldh6 | 1515/2360 | 56.02 | 4.83 |
| L-ldh7 | 1497/2182 | 54.83 | 6.14 |
| L-ldh8 | 957/1128 | 33.76 | 5.02 |
Table 2 ldh sequence characteristics of P. ostreatus
| 名称 Name | 互补DNA/基因组DNA cDNA/gDNA/bp | 分子量 Molecular weight/kD | 等电点 pI |
|---|---|---|---|
| D-ldh1 | 1473/2114 | 52.91 | 4.88 |
| D-ldh2 | 1692/2459 | 61.34 | 4.84 |
| D-ldh3 | 1776/2603 | 63.92 | 4.82 |
| L-ldh4 | 711/820 | 25.18 | 5.05 |
| L-ldh5 | 1485/2158 | 54.54 | 4.86 |
| L-ldh6 | 1515/2360 | 56.02 | 4.83 |
| L-ldh7 | 1497/2182 | 54.83 | 6.14 |
| L-ldh8 | 957/1128 | 33.76 | 5.02 |
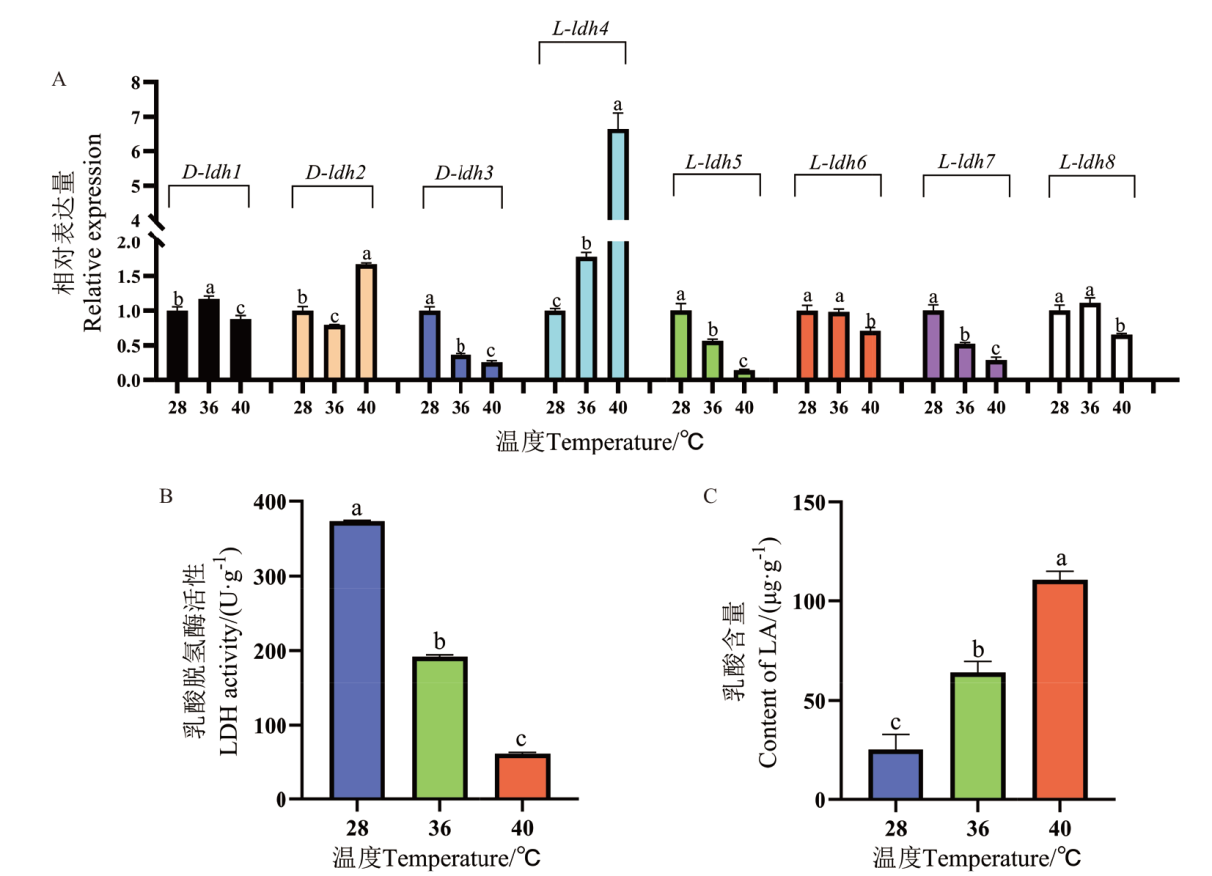
Fig. 4 Gene expression of ldh, LDH activity and LA content in P. ostreatus mycelium treated with high temperature The different letters over the columns within the same graph denote significant differences. P<0.05, the same below
| [1] |
Hong SM, Lee YK, Park I, et al. Lactic acidosis caused by repressed lactate dehydrogenase subunit B expression down-regulates mitochondrial oxidative phosphorylation via the pyruvate dehydrogenase(PDH)-PDH kinase axis[J]. Journal of Biological Chemistry, 2019, 294(19): 7810-7820.
doi: 10.1074/jbc.RA118.006095 pmid: 30923124 |
| [2] | 吴德俊. 长链非编码RNA LINK-A通过乳酸脱氢酶-A(LDH-A)促进胶质瘤细胞生长和侵袭的研究[D]. 合肥: 安徽医科大学, 2018. |
| Wu DJ. Long non-coding RNA LINK-A promotes glioma cell growth and invasion via lactate dehydrogenase A[D]. Hefei: Anhui Medical University, 2018. | |
| [3] | 安保光. 水稻D-乳酸脱氢酶基因的克隆和功能分析[D]. 武汉: 武汉大学, 2013. |
| An BG. Cloning and function analysis of a D-lactate dehydrogenase gene involved in methyglyoxal pathway in rice[D]. Wuhan: Wuhan University, 2013. | |
| [4] |
Chen WC, Wei LL, Zhang Y, et al. Involvement of the two l-lactate dehydrogenase in development and pathogenicity in Fusarium graminearum[J]. Current Genetics, 2019, 65(2): 591-605.
doi: 10.1007/s00294-018-0909-6 |
| [5] |
Zhang RY, Hu DD, Zhang YY, et al. Anoxia and anaerobic respiration are involved in “spawn-burning” syndrome for edible mushroom Pleurotus eryngii grown at high temperatures[J]. Scientia Horticulturae, 2016, 199: 75-80.
doi: 10.1016/j.scienta.2015.12.035 URL |
| [6] |
戚元成, 段庆虎, 申晓晔, 等. 高温胁迫对糙皮侧耳菌丝生理生化特性的影响[J]. 食用菌学报, 2012, 19(04): 14-16.
doi: 10.16488/j.cnki.1005-9873.2012.04.009 |
| Qi YC, Duan QH, Shen XY, et al. Selected biochemical characteristics of Pleurotus ostreatus mycelium grown at 40℃[J]. Acta Edulis Fungi, 2012, 19(04): 14-16. | |
| [7] |
Zou YJ, Zhang MJ, Qu JB, et al. iTRAQ-based quantitative proteomic analysis reveals proteomic changes in mycelium of Pleurotus ostreatus in response to heat stress and subsequent recovery[J]. Frontiers in Microbiology, 2018, 9: 2368.
doi: 10.3389/fmicb.2018.02368 URL |
| [8] |
Hou LD, Wang LN, Wu XL, et al. Expression patterns of two pal genes of Pleurotus ostreatus across developmental stages and under heat stress[J]. BMC Microbiology, 2019, 19(1): 231.
doi: 10.1186/s12866-019-1594-4 |
| [9] | 侯志浩. 糙皮侧耳Zn2Cys6转录因子家族的表达分析及PoZCP26的功能研究[D]. 北京: 中国农业科学院, 2020. |
| Hou ZH. Expression analysis of Zn2Cys6 transcription factor family and functional analysis of PoZCP26 in Pleurotus ostreatus[D]. Beijing: Chinese Academy of Agricultural Sciences Thesis. 2020. | |
| [10] |
Yan ZY, Zhao MR, Wu XL, et al. Metabolic response of Pleurotus ostreatus to continuous heat stress[J]. Frontiers in Microbiology, 2020, 10: 3148.
doi: 10.3389/fmicb.2019.03148 URL |
| [11] | 刘秀明, 黄晨阳, 等. 外源海藻糖对高温胁迫下肺形侧耳氧化损伤的缓解效应[J]. 园艺学报, 2013, 40(8): 1501-1508. |
| Liu XM, Huang CY, et al. Alleviative effects of exogenous trehalose on oxidative damage metabolism in Pleurotus pulmonarius under heat stress[J]. Acta Horticulturae Sinica, 2013, 40(8): 1501-1508. | |
| [12] | 张美敬, 刘秀明, 邹亚杰, 等. 侧耳属食用菌高温胁迫条件优化研究[J]. 菌物学报, 2015, 34(4): 662-669. |
| Zhang MJ, Liu XM, Zou YJ, et al. Optimization of heat stress for Pleurotus spp. cultivation[J]. Mycosystema, 2015, 34(4): 662-669. | |
| [13] |
Richardson RS, Noyszewski EA, Leigh JS, et al. Lactate efflux from exercising human skeletal muscle: role of intracellular Po-2[J]. Journal of Applied Physiology, 1998, 85(2): 627-634.
pmid: 9688741 |
| [14] |
Hirschhaeuser F, Sattler UGA, Mueller-Klieser W. Lactate: a metabolic Key Player in Cancer[J]. Cancer Research, 2011, 71(22): 6921-6925.
doi: 10.1158/0008-5472.CAN-11-1457 pmid: 22084445 |
| [15] |
Yang S, Wu H, He K, et al. Response of AMP-activated protein kinase and lactate metabolism of largemouth bass(Micropterus salmoides)under acute hypoxic stress[J]. Science of the Total Environment, 2019, 666: 1071-1079.
doi: 10.1016/j.scitotenv.2019.02.236 |
| [16] |
Jain M, Aggarwal S, Nagar P, et al. A D-lactate dehydrogenase from rice is involved in conferring tolerance to multiple abiotic stresses by maintaining cellular homeostasis[J]. Scientific Reports, 2020, 10(1): 12835.
doi: 10.1038/s41598-020-69742-0 pmid: 32732944 |
| [17] | 许黎明, 蒋国凤, 伍新龄, 等. 鼠李糖乳杆菌L-乳酸脱氢酶的生物信息学分析和基因克隆[J]. 食品工业科技, 2022, 1-16. |
| Xu LM, Jiang GF, et al. Bioinformatics analysis and gene cloning of L-lactate dehydrogenase from Lactobacillus rhamnosus[J]. Science and Technology of Food Industry, 2022, 1-16. | |
| [18] | Kasai T, Suzuki Y, Kouzuma A, et al. Roles of D-lactate dehydrogenases in the anaerobic growth of Shewanella oneidensis MR-1 on sugars[J]. Applied and Environmental Microbiology, 2019, 85(3): e02668-18. |
| [19] |
Bleckwedel J, Mohamed F, Mozzi F, et al. Major role of lactate dehydrogenase D-LDH1 for the synthesis of lactic acid in Fructobacillus tropaeoli CRL 2034[J]. Applied Microbiology and Biotechnology, 2020, 104(17): 7409-7426.
doi: 10.1007/s00253-020-10776-9 pmid: 32666186 |
| [20] |
Lodi T, O'Connor D, Goffrini P, et al. Carbon catabolite repression in Kluyveromyces lactis: isolation and characterization of the KIDLD gene encoding the mitochondrial enzyme D-lactate ferricytochrome c oxidoreductase[J]. Molecular and General Genetics, 1994, 244(6): 622-629.
pmid: 7969031 |
| [21] |
Engqvist M, Drincovich MF, Flugge UI, et al. Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and β-oxidation pathways[J]. Journal of Biological Chemistry, 2009, 284(37): 25026-25037.
doi: 10.1074/jbc.M109.021253 pmid: 19586914 |
| [22] | Gu SA, Jun C, Joo JC, et al. Higher thermostability of L-lactate dehydrogenases is a key factor in decreasing the optical purity of D-lactic acid produced from Lactobacillus coryniformis[J]. Enzyme and Microbial Technology, 2014, 58-59: 29-35. |
| [23] |
Sun LF, Zhang CL, Lyu PC, et al. Contributory roles of two L-lactate dehydrogenases for L-lactic acid production in thermotolerant Bacillus coagulans[J]. Scientific Reports, 2016, 6: 37916.
doi: 10.1038/srep37916 |
| [24] | 王金鹏. L-乳酸脱氢酶介导木糖葡萄球菌多重耐药机制的研究[D]. 哈尔滨: 东北农业大学, 2020. |
| Wang JP. Study of L-lactate dehydrogenase multidrug resistance mechanism on Staphylococcus xylosus[D]. Haerbin:Northeast Agricultural University, 2020. |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [3] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [4] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [5] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [6] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [7] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [8] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| [9] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [10] | REN Li, QIAO Shu-ting, GE Chen-hui, WEI Zi-tong, XU Chen-xi. Identification and Expression Analysis of Spinach PSY Gene Family [J]. Biotechnology Bulletin, 2023, 39(12): 169-178. |
| [11] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| [12] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| [13] | DUAN Min-jie, LI Yi-fei, YANG Xiao-miao, WANG Chun-ping, HUANG Qi-zhong, HUANG Ren-zhong, ZHANG Shi-cai. Identification of Zinc Finger Protein DnaJ-Like Gene Family in Capsicum annuum and Its Expression Analysis Responses to High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(1): 187-198. |
| [14] | YUAN Xing, GUO Cai-hua, LIU Jin-ming, KANG Chao, QUAN Shao-wen, NIU Jian-xin. Genome-wide Identification of CONSTANS-Like Family Genes and Expression Analysis in Wanlut [J]. Biotechnology Bulletin, 2022, 38(9): 167-179. |
| [15] | GUO Bin-hui, SONG Li. Transcription of Ethylene Biosynthesis and Signaling Associated Genes in Response to Heterodera glycine Infection [J]. Biotechnology Bulletin, 2022, 38(8): 150-158. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||