








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (11): 360-372.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0858
Previous Articles Next Articles
WANG Wen-tao( ), FENG Qi, LIU Chen-guang(
), FENG Qi, LIU Chen-guang( ), BAI Feng-wu, ZHAO Xin-qing
), BAI Feng-wu, ZHAO Xin-qing
Received:2023-09-04
Online:2023-11-26
Published:2023-12-20
Contact:
LIU Chen-guang
E-mail:tenebrae@sjtu.edu.cn;cg.liu@sjtu.edu.cn
WANG Wen-tao, FENG Qi, LIU Chen-guang, BAI Feng-wu, ZHAO Xin-qing. Redox-sensitive Genetic Parts Improve the Tolerance of Yeast to Lignocellulosic Hydrolysate Inhibitors[J]. Biotechnology Bulletin, 2023, 39(11): 360-372.
| 名称 Name | 描述 Description | 来源 Source |
|---|---|---|
| E. coli DH5α | 质粒构建 | 实验室保存 |
| S. cerevisiaeS288C | 宿主细胞 | 购买于ATCC |
| SC-PGK1 | S288C, HO-yEGFP | 实验室保存 |
| SC-TRR1 | S288C, HO-TRR1 | 本工作构建 |
| SC-TRX2 | S288C, HO-TRX2 | 本工作构建 |
| SC-MET16 | S288C, HO-MET16 | 本工作构建 |
| SC-SOD | S288C, HO-SOD | 本工作构建 |
| SC-CTT | S288C, HO-CTT | 本工作构建 |
| SC-IDP | S288C, HO-IDP | 本工作构建 |
| SC-GLR | S288C, HO-GLR | 本工作构建 |
| SC-ADH | S288C, HO-ADH | 本工作构建 |
| SC-GRE | S288C, HO-GRE | 本工作构建 |
| SC-AC | S288C, HO-AC | 本工作构建 |
Table 1 Strains used
| 名称 Name | 描述 Description | 来源 Source |
|---|---|---|
| E. coli DH5α | 质粒构建 | 实验室保存 |
| S. cerevisiaeS288C | 宿主细胞 | 购买于ATCC |
| SC-PGK1 | S288C, HO-yEGFP | 实验室保存 |
| SC-TRR1 | S288C, HO-TRR1 | 本工作构建 |
| SC-TRX2 | S288C, HO-TRX2 | 本工作构建 |
| SC-MET16 | S288C, HO-MET16 | 本工作构建 |
| SC-SOD | S288C, HO-SOD | 本工作构建 |
| SC-CTT | S288C, HO-CTT | 本工作构建 |
| SC-IDP | S288C, HO-IDP | 本工作构建 |
| SC-GLR | S288C, HO-GLR | 本工作构建 |
| SC-ADH | S288C, HO-ADH | 本工作构建 |
| SC-GRE | S288C, HO-GRE | 本工作构建 |
| SC-AC | S288C, HO-AC | 本工作构建 |
| 名称 Name | 描述 Description | 来源 Source |
|---|---|---|
| GP-SOD | PTRR1-SOD1-TADH1 | 本工作构建 |
| GP-CTT | PTRR1-CTT1-TADH1 | 本工作构建 |
| GP-IDP | PTRR1-IDP1-TADH1 | 本工作构建 |
| GP-GLR | PTRR1-GLR1-TADH1 | 本工作构建 |
| GP-ADH | PMET16-ADH6-TADH1 | 本工作构建 |
| GP-GRE | PMET16-GRE2-TADH1 | 本工作构建 |
| GP-AC | PTRR1-SOD1-TADH1- PMET16-ADH6-TADH1 | 本工作构建 |
Table 2 Genetic parts used
| 名称 Name | 描述 Description | 来源 Source |
|---|---|---|
| GP-SOD | PTRR1-SOD1-TADH1 | 本工作构建 |
| GP-CTT | PTRR1-CTT1-TADH1 | 本工作构建 |
| GP-IDP | PTRR1-IDP1-TADH1 | 本工作构建 |
| GP-GLR | PTRR1-GLR1-TADH1 | 本工作构建 |
| GP-ADH | PMET16-ADH6-TADH1 | 本工作构建 |
| GP-GRE | PMET16-GRE2-TADH1 | 本工作构建 |
| GP-AC | PTRR1-SOD1-TADH1- PMET16-ADH6-TADH1 | 本工作构建 |
| 名称 Name | 成分 Ingredient | 培养微生物 Cultured microorganism | 备注 Remark |
|---|---|---|---|
| LB | 5 g/L酵母粉、10 g/L胰蛋白胨和 10 g/L NaCl | 大肠杆菌 E. coli | 固体培养基添加20 g/L琼脂粉 |
| YPD生长 | 10 g/L酵母粉、20 g/L蛋白胨和 20 g/L葡萄糖 | 酿酒酵母 S. cerevisiae | |
| YPD发酵 | 3 g/L酵母粉、4 g/L蛋白胨和 100 g/L葡萄糖 | 酿酒酵母 S. cerevisiae |
Table 3 Names and composition of the media used
| 名称 Name | 成分 Ingredient | 培养微生物 Cultured microorganism | 备注 Remark |
|---|---|---|---|
| LB | 5 g/L酵母粉、10 g/L胰蛋白胨和 10 g/L NaCl | 大肠杆菌 E. coli | 固体培养基添加20 g/L琼脂粉 |
| YPD生长 | 10 g/L酵母粉、20 g/L蛋白胨和 20 g/L葡萄糖 | 酿酒酵母 S. cerevisiae | |
| YPD发酵 | 3 g/L酵母粉、4 g/L蛋白胨和 100 g/L葡萄糖 | 酿酒酵母 S. cerevisiae |
| 启动子 Promoter | 长度 Length/bp | GC含量 GC content/% | Yap1结合位点 Yap1 binding site |
|---|---|---|---|
| PTRR1 | 545 | 39 | 2 |
| PTRX2 | 275 | 38 | 3 |
| PMET16 | 418 | 34 | 3 |
Table 4 Sequence analysis of the promoters
| 启动子 Promoter | 长度 Length/bp | GC含量 GC content/% | Yap1结合位点 Yap1 binding site |
|---|---|---|---|
| PTRR1 | 545 | 39 | 2 |
| PTRX2 | 275 | 38 | 3 |
| PMET16 | 418 | 34 | 3 |
| 启动子 Promoter | EL | 抑制物:H2O2Inhibitor: H2O2 | 抑制物:糠醛 Inhibitor: Furfural | 抑制物:5-HMF Inhibitor: 5-HMF | |||
|---|---|---|---|---|---|---|---|
| 最大RS值 Max RS value | 对应浓度 Corresponding concentration | 最大RS值 Max RS value | 对应浓度 Corresponding concentration | 最大RS值 Max RS value | 对应浓度 Corresponding concentration | ||
| PTRR1 | 15.77 | 2.84 | 1 mmol/L | 1.64 | 10 mmol/L | 1.21 | 30 mmol/L |
| PTRX2 | 16.76 | 2.80 | 1 mmol/L | 2.40 | 10 mmol/L | 1.11 | 30 mmol/L |
| PMET16 | 1.26 | 1.27 | 1 mmol/L | 4.19 | 20 mmol/L | 1.54 | 30 mmol/L |
Table 5 Response performances of the promoters
| 启动子 Promoter | EL | 抑制物:H2O2Inhibitor: H2O2 | 抑制物:糠醛 Inhibitor: Furfural | 抑制物:5-HMF Inhibitor: 5-HMF | |||
|---|---|---|---|---|---|---|---|
| 最大RS值 Max RS value | 对应浓度 Corresponding concentration | 最大RS值 Max RS value | 对应浓度 Corresponding concentration | 最大RS值 Max RS value | 对应浓度 Corresponding concentration | ||
| PTRR1 | 15.77 | 2.84 | 1 mmol/L | 1.64 | 10 mmol/L | 1.21 | 30 mmol/L |
| PTRX2 | 16.76 | 2.80 | 1 mmol/L | 2.40 | 10 mmol/L | 1.11 | 30 mmol/L |
| PMET16 | 1.26 | 1.27 | 1 mmol/L | 4.19 | 20 mmol/L | 1.54 | 30 mmol/L |
| 菌株 Strain | 基因元件转录水平变化倍数 Fold-change in transcription level of gene element | 代谢速率变化(相比野生型) Metabolic rate change(Compared to wild type) | |||
|---|---|---|---|---|---|
| 与无胁迫比较 Compared with no stress | 胁迫下相比野生型 Compared to wild type under stress | 葡萄糖消耗速率 Glucose consumption rate/% | 乙醇生产速率 Ethanol production rate/% | 比生长速率 Specific growth rate/% | |
| SC-ADH | 3.84 | 1.12 | 27.23 | 39.99 | 20.46 |
| SC-GRE | 12.28 | 2.10 | 5.85 | 26.04 | -0.16 |
Table 9 Metabolic rate changes in recombinant strains under 5-HMF stress
| 菌株 Strain | 基因元件转录水平变化倍数 Fold-change in transcription level of gene element | 代谢速率变化(相比野生型) Metabolic rate change(Compared to wild type) | |||
|---|---|---|---|---|---|
| 与无胁迫比较 Compared with no stress | 胁迫下相比野生型 Compared to wild type under stress | 葡萄糖消耗速率 Glucose consumption rate/% | 乙醇生产速率 Ethanol production rate/% | 比生长速率 Specific growth rate/% | |
| SC-ADH | 3.84 | 1.12 | 27.23 | 39.99 | 20.46 |
| SC-GRE | 12.28 | 2.10 | 5.85 | 26.04 | -0.16 |
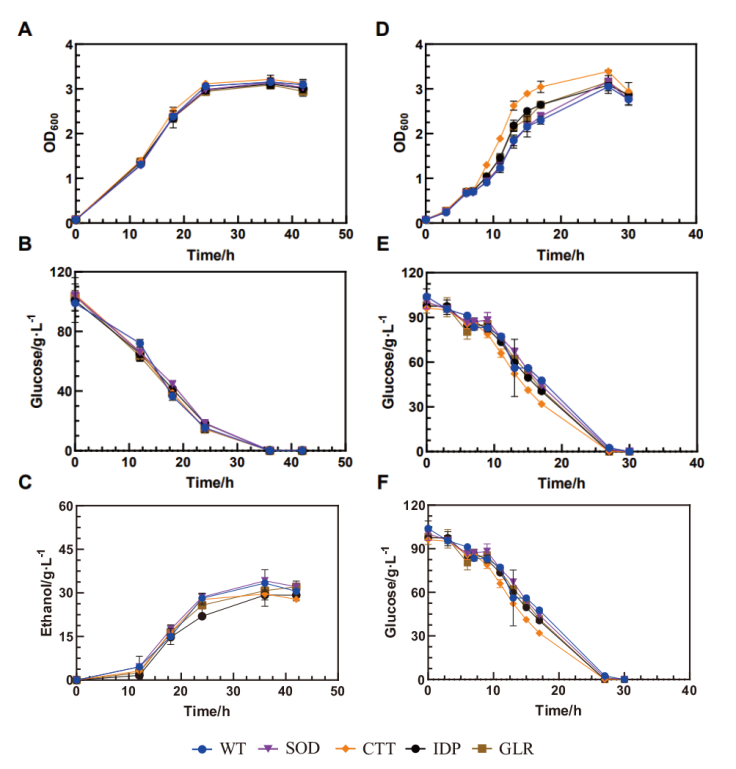
Fig. 1 Growths of the recombinant strains under stress-free and oxidative conditions A: Growth under no stress. B: Glucose consumption under no stress. C: Ethanol production under no stress. D: Growth under oxidative condition. E: Glucose consumption under oxidative condition. F: Ethanol production under oxidative condition
| 菌株 Strain | 基因元件转录水平变化倍数 Fold-change in transcription level of gene element | 代谢速率变化(相比野生型) Metabolic rate change(Compared to wild type) | |||||
|---|---|---|---|---|---|---|---|
| 与无胁迫比较 Compared with no stress | 胁迫下相比野生型 Compared to wild type under stress | 葡萄糖消耗速率 Glucose consumption rate | 乙醇生产速率 Ethanol production rate | 比生长速率 Specific growth rate | |||
| SC-CTT | 2.42 | 130 | 30.23% | 32.79% | 32.51% | ||
| SC-SOD | 4.34 | 5.33 | 6.47% | 4.37% | 3.79% | ||
| SC-IDP | 1.34 | 1.13 | 13.68% | 12.02% | 15.12% | ||
| SC-GLR | 1.57 | 6.56 | 11.67% | 16.34% | 15.03% | ||
Table 6 Transcriptional level changes of genetic parts and metabolic rate changes in recombinant strains
| 菌株 Strain | 基因元件转录水平变化倍数 Fold-change in transcription level of gene element | 代谢速率变化(相比野生型) Metabolic rate change(Compared to wild type) | |||||
|---|---|---|---|---|---|---|---|
| 与无胁迫比较 Compared with no stress | 胁迫下相比野生型 Compared to wild type under stress | 葡萄糖消耗速率 Glucose consumption rate | 乙醇生产速率 Ethanol production rate | 比生长速率 Specific growth rate | |||
| SC-CTT | 2.42 | 130 | 30.23% | 32.79% | 32.51% | ||
| SC-SOD | 4.34 | 5.33 | 6.47% | 4.37% | 3.79% | ||
| SC-IDP | 1.34 | 1.13 | 13.68% | 12.02% | 15.12% | ||
| SC-GLR | 1.57 | 6.56 | 11.67% | 16.34% | 15.03% | ||
| 菌株 Strain | ROS荧光强度 ROS fluorescence intensity |
|---|---|
| WT | 2017±373 |
| SC-CTT | 1221±48 |
| SC-SOD | 1401±185 |
| SC-IDP | 1428±66 |
| SC-GLR | 1465±104 |
Table 7 ROS levels in strains
| 菌株 Strain | ROS荧光强度 ROS fluorescence intensity |
|---|---|
| WT | 2017±373 |
| SC-CTT | 1221±48 |
| SC-SOD | 1401±185 |
| SC-IDP | 1428±66 |
| SC-GLR | 1465±104 |
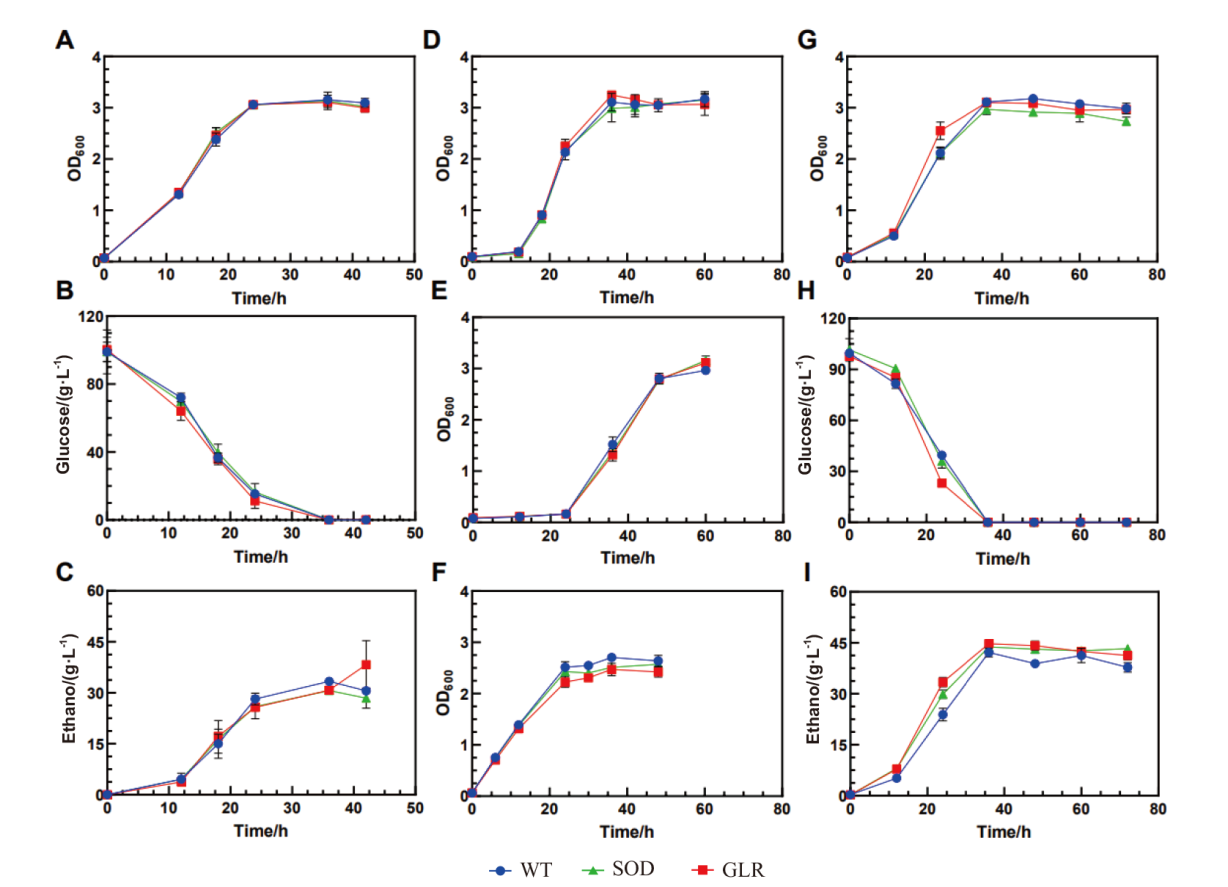
Fig. 2 Growths of the recombinant strains under stress-free, furfural stress and 5-HMF stress conditions A: Growth under no stress. B: Glucose consumption under no stress. C: Ethanol production under no stress. D: Growth curve under 3 g/L furfural stress. E: Growth curve under 4 g/L furfural stress. F: Shock experiment with 4 g/L furfural. G: Growth under 5-HMF stress. H: Glucose consumption under 5-HMF stress. I: Ethanol production under 5-HMF stress
| 菌株 Strain | 糠醛胁迫下ROS荧光强度 ROS fluorescence intensity under furfural stress | 5-HMF胁迫下ROS荧光强度 ROS fluorescence intensity under 5-HMF stress |
|---|---|---|
| WT | 1118±219 | 1281±395 |
| SC-ADH | 1200±182 | 913±23 |
| SC-GRE | 1618±149 | 1135±203 |
Table 8 ROS levels under furfural stress and 5-HMF stress
| 菌株 Strain | 糠醛胁迫下ROS荧光强度 ROS fluorescence intensity under furfural stress | 5-HMF胁迫下ROS荧光强度 ROS fluorescence intensity under 5-HMF stress |
|---|---|---|
| WT | 1118±219 | 1281±395 |
| SC-ADH | 1200±182 | 913±23 |
| SC-GRE | 1618±149 | 1135±203 |
| 菌株 Strain | 添加抑制物时期 Inhibitor addition period | 葡萄糖消耗速率 Glucose consumption rate/% | 乙醇生产速率 Ethanol production rate/% | 比生长速率 Specific growth rate/% |
|---|---|---|---|---|
| SC-ADH | 无胁迫 | 1.4 | 4.6 | 3.3 |
| 对数期 | 4.63 | 1.16 | 4.62 | |
| 稳定期 | 3.08 | 5.90 | 0.25 | |
| SC-CTT | 无胁迫 | 3.4 | -0.1 | 1.1 |
| 对数期 | 17.4 | 5.68 | 17.8 | |
| 稳定期 | 1.07 | 3.10 | 1.68 | |
| SC-AC | 无胁迫 | 6.7 | -0.8 | 0.7 |
| 对数期 | 31.0 | 41.7 | 24.3 | |
| 稳定期 | 3.07 | 4.86 | 1.26 |
Table 10 Performance improvements of dual genetic parts in response to complex stress(Compared to the wild-type)
| 菌株 Strain | 添加抑制物时期 Inhibitor addition period | 葡萄糖消耗速率 Glucose consumption rate/% | 乙醇生产速率 Ethanol production rate/% | 比生长速率 Specific growth rate/% |
|---|---|---|---|---|
| SC-ADH | 无胁迫 | 1.4 | 4.6 | 3.3 |
| 对数期 | 4.63 | 1.16 | 4.62 | |
| 稳定期 | 3.08 | 5.90 | 0.25 | |
| SC-CTT | 无胁迫 | 3.4 | -0.1 | 1.1 |
| 对数期 | 17.4 | 5.68 | 17.8 | |
| 稳定期 | 1.07 | 3.10 | 1.68 | |
| SC-AC | 无胁迫 | 6.7 | -0.8 | 0.7 |
| 对数期 | 31.0 | 41.7 | 24.3 | |
| 稳定期 | 3.07 | 4.86 | 1.26 |
| 菌株 Strain | 总生物量 Total biomass | 葡萄糖消耗速率 Glucose consumption rate | 乙醇生产速率 Ethanol production rate | 比生长速率 Specific growth rate |
|---|---|---|---|---|
| SC-AC | 11.5% | 60.1% | 58.9% | 64.2% |
Table 11 Performance improvements of dual genetic parts in response to complex stress(Adding inhibitors during the lag phase, compared to SC-CTT strain)
| 菌株 Strain | 总生物量 Total biomass | 葡萄糖消耗速率 Glucose consumption rate | 乙醇生产速率 Ethanol production rate | 比生长速率 Specific growth rate |
|---|---|---|---|---|
| SC-AC | 11.5% | 60.1% | 58.9% | 64.2% |
| 菌株 Strain | ROS level | NAD+/ NADH | NADP+/ NADPH | ATP content/ (nmol·mg-1Protein) | Total GSH content/(mmol·mg-1Protein) | SOD enzyme activity/(U·mg-1Protein) | CAT enzyme activity/(U·mg-1Protein) |
|---|---|---|---|---|---|---|---|
| SC-CTT | 324±5 | 0.32±0.04 | 0.26±0.02 | 11.54±0.33 | 16.98±3.29 | 295±15 | 26.10±1.22 |
| SC-AC | 312±2 | 0.30±0.14 | 0.28±0.01 | 8.34±0.52 | 23.37±4.45 | 242±98 | 36.60±0.09 |
Table 12 Characterization of the physiological state of the recombinant strains in the lag phase stress group
| 菌株 Strain | ROS level | NAD+/ NADH | NADP+/ NADPH | ATP content/ (nmol·mg-1Protein) | Total GSH content/(mmol·mg-1Protein) | SOD enzyme activity/(U·mg-1Protein) | CAT enzyme activity/(U·mg-1Protein) |
|---|---|---|---|---|---|---|---|
| SC-CTT | 324±5 | 0.32±0.04 | 0.26±0.02 | 11.54±0.33 | 16.98±3.29 | 295±15 | 26.10±1.22 |
| SC-AC | 312±2 | 0.30±0.14 | 0.28±0.01 | 8.34±0.52 | 23.37±4.45 | 242±98 | 36.60±0.09 |
| [1] |
Rittmann BE. Opportunities for renewable bioenergy using microorganisms[J]. Biotechnology and Bioengineering, 2008, 100(2):203-212.
doi: 10.1002/bit.21875 pmid: 18431744 |
| [2] |
Zhang J, Wang YH, Du XJ, et al. Selective removal of lignin to enhance the process of preparing fermentable sugars and platform chemicals from lignocellulosic biomass[J]. Bioresource Technology, 2020, 303:122846.
doi: 10.1016/j.biortech.2020.122846 URL |
| [3] |
Zhang ZR, Song JL, Han BX. Catalytic transformation of lignocellulose into chemicals and fuel products in ionic liquids[J]. Chemical Reviews, 2017, 117(10):6834-6880.
doi: 10.1021/acs.chemrev.6b00457 pmid: 28535680 |
| [4] |
Chen CY, Zhao XQ, Yen HW, et al. Microalgae-based carbohydrates for biofuel production[J]. Biochemical Engineering Journal, 2013, 78:1-10.
doi: 10.1016/j.bej.2013.03.006 URL |
| [5] |
Zhao W, Zhao F, Zhang S, et al. Ethanol production by simultaneous saccharification and cofermentation of pretreated corn stalk[J]. Journal of Basic Microbiology, 2019, 59(7):744-753.
doi: 10.1002/jobm.201900117 pmid: 31087563 |
| [6] | Robak K, Balcerek M. Current state-of-the-art in ethanol production from lignocellulosic feedstocks[J]. Microbiology Research, 2020, 240:126534. |
| [7] | Deshavath NN, Mohan M, Veeranki VD, et al. Dilute acid pretreatment of sorghum biomass to maximize the hemicellulose hydrolysis with minimized levels of fermentative inhibitors for bioethanol production[J]. Biotechnology, 2017, 7(2):139. |
| [8] |
Allen SA, Clark W, Mccaffery JM, et al. Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae[J]. Biotechnology for Biofuels, 2010, 3(1):2.
doi: 10.1186/1754-6834-3-2 |
| [9] |
Du X, Takagi H. N-Acetyltransferase Mpr1 confers ethanol tolerance on Saccharomyces cerevisiae by reducing reactive oxygen species[J]. Applied Microbiology and Biotechnology, 2007, 75(6):1343-1351.
doi: 10.1007/s00253-007-0940-x URL |
| [10] |
Herrero E, Ros J, Bellí G, et al. Redox control and oxidative stress in yeast cells[J]. Biochimica et Biophysica Acta, 2008, 1780(11):1217-1235.
doi: 10.1016/j.bbagen.2007.12.004 pmid: 18178164 |
| [11] | 马超颖, 戚薇, 杜连祥. 酿酒酵母氧化应激反应的研究进展[J]. 天津轻工业学院学报, 2002, 4: 14-17+31. |
| Ma CY, Qi W, Du LX. Research progress on oxidative stress response of Saccharomyces cerevisiae[J]. Journal of Tianjin University of Science and Technology, 2002, 4: 14-17+31. | |
| [12] |
Jayakody LN, Jin YS. In-depth understanding of molecular mechanisms of aldehyde toxicity to engineer robust Saccharomyces cerevisiae[J]. Applied Microbiology and Biotechnology, 2021, 105(7):2675-2692.
doi: 10.1007/s00253-021-11213-1 pmid: 33743026 |
| [13] | 杨雪雪, 蒋伶活. 酵母对木质纤维素酸解物中抑制物的应答及菌株开发[J]. 生命科学, 2012, 24(6):531-534. |
| Yang XX, Jiang LH. The response of yeast to inhibitors in lignocellulosic acidolysis and strain development[J]. Life Science, 2012, 24(6):531-534. | |
| [14] |
Vemuri GN, Altman E, Sangurdekar DP, et al. Overflow metabolism in Escherichia coli during steady-state growth: Transcriptional regulation and effect of the redox ratio[J]. Applied and Environmental Microbiology, 72(5):3653-3661.
doi: 10.1128/AEM.72.5.3653-3661.2006 URL |
| [15] |
Mason JT, Kim SK, Knaff DB, et al. Thermodynamic basis for redox regulation of the Yap1 signal transduction pathway[J]. Biochemistry, 2006, 45(45):13409-13417.
pmid: 17087494 |
| [16] |
Liu CG, Lin YH, Bai FW. Global gene expression analysis of Saccharomyces cerevisiae grown under redox potential controlled very-high-gravity conditions[J]. Biotechnology Journal, 2014, 8(11):1332-1340.
doi: 10.1002/biot.v8.11 URL |
| [17] |
Liu CG, Xue C, Lin YH, et al. Redox potential control and applications in microaerobic and anaerobic fermentations[J]. Biotechnology Advances, 2013, 31(2):257-265.
doi: 10.1016/j.biotechadv.2012.11.005 URL |
| [18] |
De La Torre-Ruiz MA, Pujol N, Sundaran V. Coping with oxidative stress. The yeast model[J]. Current Drug Targets, 2015, 16(1):2-12.
pmid: 25330032 |
| [19] | Hao X, Du B, Liu L, et al. Effect of ORP regulation on yeast fermentation with inhibitors of lignocellulose hydrolysate[J]. Ciesc Journal, 2015, 66(3):1066-1071. |
| [20] |
Li K, Xia J, Mehmood MA, et al. Extracellular redox potential regulation improves yeast tolerance to furfural[J]. Chemical Engineering Science, 2019, 196:54-63.
doi: 10.1016/j.ces.2018.11.059 URL |
| [21] |
Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method[J]. Nature Protocols, 2007, 2(1):31-34.
doi: 10.1038/nprot.2007.13 pmid: 17401334 |
| [22] |
Teixeira MC, Monteiro PT, Sa-Correia I. Predicting gene and genomic regulation in Saccharomyces cerevisiae, using the YEASTRACT Database: A step-by-step guided analysis[J]. Methods in Molecular Biology, 2016, 1361:391-404.
doi: 10.1007/978-1-4939-3079-1_22 pmid: 26483034 |
| [23] | 刘广新, 刘玲彦, 王林, 等. 啤酒酵母活性/活力检测方法比较及其与发酵性能的相关性研究[J]. 中外酒业, 2020, 9:40-54. |
| Liu GX, Liu LY, Wang L, et al. Comparison of assay methods for activity/vitality of brewer's yeast and its correlation with fermentation performance[J]. Chinese and foreign wine industry, 2020, 9:40-54. | |
| [24] |
Lee J, Godon C, Lagniel G, et al. Yap1 and Skn7 Control two specialized oxidative stress response regulons in yeast[J]. Journal of Biological Chemistry, 1999, 274(23):16040-16046.
doi: 10.1074/jbc.274.23.16040 pmid: 10347154 |
| [25] |
Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides[J]. Embo Journal, 1994, 13(3):655-664
doi: 10.1002/j.1460-2075.1994.tb06304.x pmid: 8313910 |
| [26] |
Ouyang X, Tran QT, Goodwin S, et al. Yap1 activation by H2O2 or thiol-reactive chemicals elicits distinct adaptive gene responses[J]. Free Radical Biology and Medicine, 2011, 50(1):1-13.
doi: 10.1016/j.freeradbiomed.2010.10.697 pmid: 20971184 |
| [27] |
Thomas D, Barbey R, Surdin-Kerjan Y. Gene-enzyme relationship in the sulfate assimilation pathway of Saccharomyces cerevisiae. Study of the 3'-phosphoadenylylsulfate reductase structural gene[J]. Journal of Biological Chemistry, 1990, 265(26):15518-15524.
pmid: 2203779 |
| [28] |
Kim D, Hahn JS. Roles of the Yap1 transcription factor and antioxidants in Saccharomyces cerevisiae's tolerance to furfural and 5-hydroxymethylfurfural, which function as thiol-reactive electrophiles generating oxidative stress[J]. Applied and Environmental Microbiology, 2013, 79(16):5069-5077.
doi: 10.1128/AEM.00643-13 URL |
| [29] |
Benjaphokee S, Hasegawa D, Yokota D, et al. Highly efficient bioethanol production by a Saccharomyces cerevisiae strain with multiple stress tolerance to high temperature, acid and ethanol[J]. Nature Biotechnology, 2012, 29(3):379-386.
doi: 10.1038/nbt0511-379 |
| [30] |
Qiu Z, Jiang R. Improving Saccharomyces cerevisiae ethanol production and tolerance via RNA polymerase II subunit Rpb7[J]. Biotechnology for Biofuels, 2017, 10:125.
doi: 10.1186/s13068-017-0806-0 URL |
| [31] |
Gechev TS, Van Breusegem F, Stone JM, et al. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death[J]. Bioessays, 2006, 28(11):1091-1101.
doi: 10.1002/bies.20493 pmid: 17041898 |
| [32] |
Azad GK, Singh V, Tomar RS. Assessment of the biological pathways targeted by isocyanate using N-succinimidyl N-methylcarbamate in budding yeast Saccharomyces cerevisiae[J]. PLoS One, 2014, 9(3):e92993.
doi: 10.1371/journal.pone.0092993 URL |
| [33] | Nishimoto T, Furuta M, Kataoka M, et al. Important role of catalase in the cellular response of the budding yeast Saccharomyces cerevisiae exposed to ionizing radiation[J]. Currunt Microbiology, 2015, 70(3):404-407. |
| [34] |
Qin J, Zhou YJ, Krivoruchko A, et al. Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine[J]. Nature Communications, 2015, 6:8224.
doi: 10.1038/ncomms9224 |
| [35] |
Ask M, Mapelli V, Höck H, et al. Engineering glutathione biosynthesis of Saccharomyces cerevisiae increases robustness to inhibitors in pretreated lignocellulosic materials[J]. Microbial Cell Factories, 2013, 12(1):87-87.
doi: 10.1186/1475-2859-12-87 |
| [36] |
Outten CE, Culotta VC. Alternative start sites in the Saccharomyces cerevisiae GLR1 gene are responsible for mitochondrial and cytosolic isoforms of glutathione reductase[J]. Journal of Biological Chemistry, 2004, 279(9):7785-7791.
doi: 10.1074/jbc.M312421200 pmid: 14672937 |
| [37] |
Daehwan K. Physico-chemical conversion of lignocellulose inhibitor effects and detoxification strategies: A mini review[J]. Molecules, 2018, 23(2):309-.
doi: 10.3390/molecules23020309 URL |
| [38] | Larroy C, Parés X, Biosca JA. Characterization of a Saccharomyces cerevisiae NADP(H)-dependent alcohol dehydrogenase(ADHVII), a member of the cinnamyl alcohol dehydrogenase family[J]. FEBS Journal, 2002, 269(22):5738-5745. |
| [39] |
Petersson A, Almeida JR, Modig T, et al. A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance[J]. Yeast, 2006, 23(6):455-464.
doi: 10.1002/yea.1370 pmid: 16652391 |
| [40] |
Moon J, Liu ZL. Engineered NADH-dependent GRE2 from Saccharomyces cerevisiae by directed enzyme evolution enhances HMF reduction using additional cofactor NADPH[J]. Enzyme and Microbial Technology, 2012, 50(2):115-120.
doi: 10.1016/j.enzmictec.2011.10.007 URL |
| [1] | XU Fa-di, XU Kang, SUN Dong-ming, LI Meng-lei, ZHAO Jian-zhi, BAO Xiao-ming. Research Progress in Second-generation Fuel Ethanol Technology Based on Poplar(Populus sp.) [J]. Biotechnology Bulletin, 2023, 39(9): 27-39. |
| [2] | CHENG Ting, YUAN Shuai, ZHANG Xiao-yuan, LIN Liang-cai, LI Xin, ZHANG Cui-ying. Research Progress in the Regulation of Isobutanol Synthesis Pathway in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(7): 80-90. |
| [3] | LI Xin-yue, ZHOU Ming-hai, FAN Ya-chao, LIAO Sha, ZHANG Feng-li, LIU Chen-guang, SUN Yue, ZHANG Lin, ZHAO Xin-qing. Research Progress in the Improvement of Microbial Strain Tolerance and Efficiency of Biological Manufacturing Based on Transporter Engineering [J]. Biotechnology Bulletin, 2023, 39(11): 123-136. |
| [4] | ZHU Ying-xuan, LI Ke-jing, HE Min, ZHENG Dao-qiong. Research Progress in the Exploring Genomic Variations Driven by Stress Factors Using the Yeast Model [J]. Biotechnology Bulletin, 2023, 39(11): 191-204. |
| [5] | TANG Rui-qi, ZHAO Xin-qing, ZHU Du, WANG Ya. Stress Tolerance of Escherichia coli to Inhibitors in Lignocellulosic Hydrolysates [J]. Biotechnology Bulletin, 2023, 39(11): 205-216. |
| [6] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [7] | CHEN Hong-yan, LI Xiao-er, LI Zhong-guang. Sugar Signaling and Its Role in Plant Response to Environmental Stress [J]. Biotechnology Bulletin, 2022, 38(7): 80-89. |
| [8] | ZHAI Xu-hang, LI Xia, YUAN Ying-jin. Research Progress of Lignocellulose Pretreatment and Valorization Method [J]. Biotechnology Bulletin, 2021, 37(3): 162-174. |
| [9] | HU Fang, DONG Xu, SHI Chang-wei, WU Xue-dong. Progress in Ultrasound Intensification for Enzymatic Hydrolysis of Lignocellulose [J]. Biotechnology Bulletin, 2021, 37(10): 234-244. |
| [10] | CUI Xin-gang, SUN Ya-xin, CUI Xiao-jing, DENG Yan-wen, SUN En-hao, WANG Jun-fang, CUI Hong-jing. Roles of Gene TAP42 in the Cell Wall Stress Response of Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2021, 37(10): 57-62. |
| [11] | GU Han-qi, SHAO Ling-zhi, LIU Ran, LIU Xiao-guang, LI Ling, LIU Qian, LI Jie, ZHANG Ya-li. Lipidomics Analysis of Saccharomyces cerevisiae with Tolerance to Phenolic Inhibitors [J]. Biotechnology Bulletin, 2021, 37(1): 15-23. |
| [12] | GUO Zhen-qiang, ZHANG Yong, CAO Yun-qi, LIU Yun-yun, ZHAO Yu, WU Ai-min. Research Progress of Fuel Ethanol Fermentation Technology [J]. Biotechnology Bulletin, 2020, 36(1): 238-244. |
| [13] | ZHANG Jia-shun, GAO Li-li, MA Jiang-shan, LIU Gao-qiang. Effect of Surfactant on Cellulase Hydrolysis and Its Mechanism [J]. Biotechnology Bulletin, 2019, 35(9): 11-20. |
| [14] | ZHANG Dan, WANG Nan, LI Chao, XIE Qi, TANG San-yuan. Sweet Sorghum—a High Efficient and Quality Forage Crop [J]. Biotechnology Bulletin, 2019, 35(5): 2-8. |
| [15] | CAO Yun-qi, LIU Yun-yun, HU Nan-jiang, HU Xiao-wei, ZHANG Yao, ZHAO Yu, WU Ai-min. Current Status and Prospects of Fuel Ethanol Production [J]. Biotechnology Bulletin, 2019, 35(4): 163-169. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||