








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (4): 159-166.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0962
Previous Articles Next Articles
XIAO Ya-ru( ), JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia(
), JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia( )
)
Received:2023-10-11
Online:2024-04-26
Published:2024-04-30
Contact:
LI Li-xia
E-mail:cucumberteam2021@126.com;lilixia222@126.com
XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber[J]. Biotechnology Bulletin, 2024, 40(4): 159-166.
| 用途 Application | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| 实时荧光定量PCR RT-qPCR | TUA-F | AGCGTTTGTCTGTTGACTATGGA |
| TUA-R | TGCAGATATCATAAATGGCTTCGT | |
| CsERF025L-qF | GGTTGCAATGCCGTTCTCAA | |
| CsERF025L-qR | ACAGCGCTTCTTCATCGACA | |
| 基因克隆 Gene cloning | CsERF025L-F | ATGGCCTCTACATCCTCCAAGAG |
| CsERF025L-R | CTAGTGGTAGCTCCAAAGATTTCCAT |
Table 1 Primers for CsERF025L transcription factor cloning and real-time fluorescence quantification
| 用途 Application | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| 实时荧光定量PCR RT-qPCR | TUA-F | AGCGTTTGTCTGTTGACTATGGA |
| TUA-R | TGCAGATATCATAAATGGCTTCGT | |
| CsERF025L-qF | GGTTGCAATGCCGTTCTCAA | |
| CsERF025L-qR | ACAGCGCTTCTTCATCGACA | |
| 基因克隆 Gene cloning | CsERF025L-F | ATGGCCTCTACATCCTCCAAGAG |
| CsERF025L-R | CTAGTGGTAGCTCCAAAGATTTCCAT |
| 基因名称 Gene name | 氨基酸数 NAA | 分子质量 Mw/kD | 等电点 pI | 不稳定系数 II | 亲水系数 GRAVY | 带负电荷的残基总数 Asp+Glu | 带正电荷的残基总数 Arg+Lys | 脂肪族指数 AI | 亚细胞定位 SL |
|---|---|---|---|---|---|---|---|---|---|
| CsERF025L | 169 | 1.87 | 6.64 | 57.22 | -0.620 | 17 | 16 | 58.46 | 细胞核 |
Table 2 Physicochemical properties of proteins in cucumber CsERF025L
| 基因名称 Gene name | 氨基酸数 NAA | 分子质量 Mw/kD | 等电点 pI | 不稳定系数 II | 亲水系数 GRAVY | 带负电荷的残基总数 Asp+Glu | 带正电荷的残基总数 Arg+Lys | 脂肪族指数 AI | 亚细胞定位 SL |
|---|---|---|---|---|---|---|---|---|---|
| CsERF025L | 169 | 1.87 | 6.64 | 57.22 | -0.620 | 17 | 16 | 58.46 | 细胞核 |

Fig. 2 Protein analysis of cucumber CsERF025L A: Secondary structure of cucumber CsERF025L. B: Tertiary structure of cucumber CsERF025L protein. C: Prediction map of cucumber CsERF025L phosphorylation. D: Protein interaction network model of cucumber CsERF025L
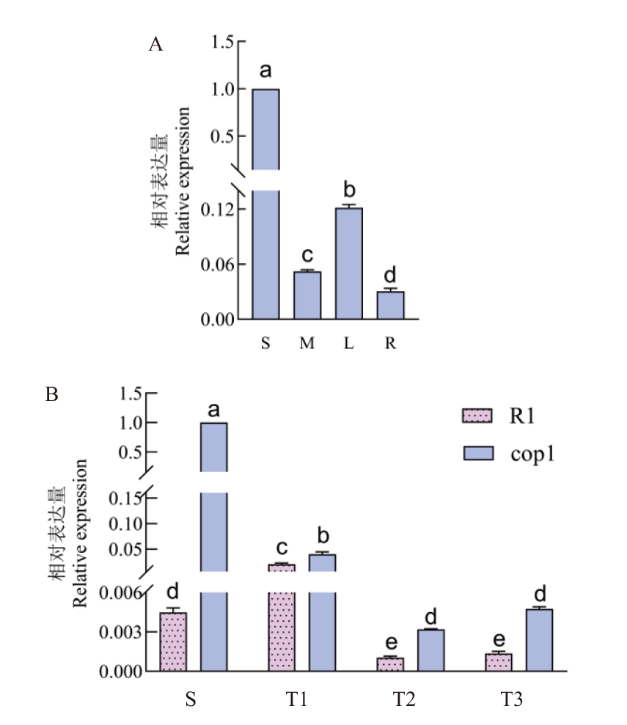
Fig. 6 Specific expression of CsERF025L in cucumber S: Seed(the first day after soaking); M: stem; L: leaf; R: root; T1: stem tip 1(the seventh day after soaking); T2: stem tip 2(the 12th day after soaking; T3: stem tip 3(the 17th day after soaking); R1: alternate phyllotaxis in cucumber; cop1: opposite phyllotaxis in cucumber; Different lowercases indicate significant differences at the 0.05 level
| [4] |
Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors[J]. Biol Chem, 1998, 379(6): 633-646.
doi: 10.1515/bchm.1998.379.6.633 pmid: 9687012 |
| [5] | Gu C, Guo ZH, Hao PP, et al. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm[J]. Bot Stud, 2017, 58(1): 6. |
| [6] |
Gao Y, Han D, Jia W, et al. Molecular characterization and systematic analysis of NtAP2/ERF in tobacco and functional determination of NtRAV-4 under drought stress[J]. Plant Physiol Biochem, 2020, 156: 420-435.
doi: 10.1016/j.plaphy.2020.09.027 URL |
| [7] |
Nakano T, Suzuki K, Fujimura T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice[J]. Plant Physiol, 2006, 140(2): 411-432.
doi: 10.1104/pp.105.073783 URL |
| [8] |
潘健, 温海帆, 何欢乐, 等. 黄瓜ERF基因家族鉴定及其在雌花芽分化中的表达分析[J]. 中国农业科学, 2020, 53(1): 133-147.
doi: 10.3864/j.issn.0578-1752.2020.01.013 |
| Pan J, Wen HF, He HL, et al. Genome-wide identification of cucumber ERF gene family and expression analysis in female bud differentiation[J]. Sci Agric Sin, 2020, 53(1): 133-147. | |
| [9] |
Hu LF, Liu SQ. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers[J]. Genet Mol Biol, 2011, 34(4): 624-633.
doi: 10.1590/S1415-47572011005000054 URL |
| [10] | Sekhar S, Kumar J, Mohanty S, et al. Identification of novel QTLs for grain fertility and associated traits to decipher poor grain filling of basal spikelets in dense panicle rice[J]. Sci Rep, 2021, 11(1): 13617. |
| [11] | Xu Y, Li XN, Yang X, et al. Genome-wide identification and molecular characterization of the AP2/ERF superfamily members in sand pear(Pyrus pyrifolia)[J]. BMC Genomics, 2023, 24(1): 32. |
| [12] |
Lai PH, Huang LM, Pan ZJ, et al. PeERF1, a SHINE-like transcription factor, is involved in nanoridge development on lip epidermis of Phalaenopsis flowers[J]. Front Plant Sci, 2020, 10: 1709.
doi: 10.3389/fpls.2019.01709 URL |
| [13] |
Saelim L, Akiyoshi N, Tan TT, et al. Arabidopsis Group IIId ERF proteins positively regulate primary cell wall-type CESA genes[J]. J Plant Res, 2019, 132(1): 117-129.
doi: 10.1007/s10265-018-1074-1 |
| [14] |
Zhang M, Li ST, Nie L, et al. Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis[J]. Plant Mol Biol, 2015, 89(4-5): 463-473.
doi: 10.1007/s11103-015-0382-2 pmid: 26445975 |
| [15] | 崔馨文, 王燕燕. 喜树转录因子CaERF1原核表达及蛋白纯化[J/OL]. 分子植物育种, 2022. https://kns.cnki.net/kcms/detail/46.1068.S.20221015.1047.002.html. |
| Cui XW, Wang YY. Prokaryotic expression and purification of transcription factor CaERF1 from Camptotheca acuminata Decne[J/OL]. Molecular Plant Breeding, 2022. https://kns.cnki.net/kcms/detail/46.1068.S.20221015.1047.002.html. | |
| [16] | Yu F, Tan ZD, Fang T, et al. A comprehensive transcriptomics analysis reveals long non-coding RNA to be involved in the key metabolic pathway in response to waterlogging stress in maize[J]. Genes, 2020, 11(3): 267. |
| [17] | Park SI, Kwon HJ, et al. The OsERF115/AP2EREBP110 transcription factor is involved in the multiple stress tolerance to heat and drought in rice plants[J]. Int J Mol Sci, 2021, 22(13): 7181. |
| [18] | Xu Y, Lu JH, Zhang JD, et al. Transcriptome revealed the molecular mechanism of Glycyrrhiza inflata root to maintain growth and development, absorb and distribute ions under salt stress[J]. BMC Plant Biol, 2021, 21(1): 599. |
| [19] |
Sandhu D, Pudussery MV, et al. Characterization of natural genetic variation identifies multiple genes involved in salt tolerance in maize[J]. Funct Integr Genomics, 2020, 20(2): 261-275.
doi: 10.1007/s10142-019-00707-x |
| [20] |
Xie ZY, Wang J, Wang WS, et al. Integrated analysis of the transcriptome and metabolome revealed the molecular mechanisms underlying the enhanced salt tolerance of rice due to the application of exogenous melatonin[J]. Front Plant Sci, 2021, 11: 618680.
doi: 10.3389/fpls.2020.618680 URL |
| [21] | 徐乐, 陈培茹, 刘意, 等. 甘薯IbERF071基因的克隆及表达特性分析[J]. 长江大学学报: 自然科学版, 2022, 19(3): 99-108. |
| Xu L, Chen PR, Liu Y, et al. Cloning and expression pattern analysis of IbERF071 gene from sweet potato[J]. J Yangtze Univ Nat Sci Ed, 2022, 19(3): 99-108. | |
| [22] |
Tahmasebi A, Khahani B, Tavakol E, et al. Microarray analysis of Arabidopsis thaliana exposed to single and mixed infections with Cucumber mosaic virus and turnip viruses[J]. Physiol Mol Biol Plants, 2021, 27(1): 11-27.
doi: 10.1007/s12298-021-00925-3 |
| [23] |
Tezuka D, Kawamata A, et al. The rice ethylene response factor OsERF83 positively regulates disease resistance to Magnaporthe oryzae[J]. Plant Physiol Biochem, 2019, 135: 263-271.
doi: 10.1016/j.plaphy.2018.12.017 URL |
| [24] |
Gebretsadik K, Qiu XY, Dong SY, et al. Molecular research progress and improvement approach of fruit quality traits in cucumber[J]. Theor Appl Genet, 2021, 134(11): 3535-3552.
doi: 10.1007/s00122-021-03895-y pmid: 34181057 |
| [25] | Gou CX, Zhu PY, Meng YJ, et al. Evaluation and genetic analysis of parthenocarpic germplasms in cucumber[J]. Genes, 2022, 13(2): 225. |
| [26] | 刘坤. 中间锦鸡儿瞬时表达体系的建立及AP2/ERF基因家族的鉴定、分析和功能研究[D]. 呼和浩特: 内蒙古农业大学, 2018. |
| Liu K. Establishment of transient expression system and identification, analysis and functional study of AP2/ERF gene family from Caragana intermedia[D]. Hohhot: Inner Mongolia Agricultural University, 2018. | |
| [27] | Tang YS, Zhang GR, Jiang XY, et al. Genome-wide association study of glucosinolate metabolites(mGWAS)in Brassica napus L[J]. Plants, 2023, 12(3): 639. |
| [28] |
Wang CH, Xin M, Zhou XY, et al. The novel ethylene-responsive factor CsERF025 affects the development of fruit bending in cucumber[J]. Plant Mol Biol, 2017, 95(4-5): 519-531.
doi: 10.1007/s11103-017-0671-z pmid: 29052099 |
| [29] | Mann R, Notani D. Transcription factor condensates and signaling driven transcription[J]. Nucleus, 2023, 14(1): 2205758. |
| [30] | 张通. bHLH类转录因子RhBEE3通过促进ABA合成调节月季花朵衰老的机理研究[D]. 武汉: 华中农业大学, 2021. |
| Zhang T. The mechanism of bHLH transcription factor RhBEE3 mediates rose flower senescence by promoting ABA synthesis[D]. Wuhan: Huazhong Agricultural University, 2021. | |
| [31] | 肖亮. AP2/ERF转录因子LTF1调控菘蓝活性木脂素生物合成的机制研究[D]. 上海: 中国人民解放军海军军医大学, 2021. |
| Xiao L. Study on the mechanism of AP2/ERF transcription factor LTF1 regulating lignan biosynthesis in Isatis indigotica[D]. Shanghai: Naval Medical University, 2021. | |
| [32] | 李瑞熙, 李洁如, 瞿礼嘉. 拟南芥MATE家族基因ADP1通过调节生长素水平调控植物株型建成[C]// 2011全国植物生物学研讨会论文集. 南宁, 2011: 33. |
| Li RX, Li JR, Qu LJ. Arabidopsis MATE family gene ADP1 regulates plant formation by regulating auxin levels[C]// 2011Proceedings of the National Symposium on Plant Biology. Nanning, 2011: 33. | |
| [33] |
Scheres B, Krizek BA. Coordination of growth in root and shoot apices by AIL/PLT transcription factors[J]. Curr Opin Plant Biol, 2018, 41: 95-101.
doi: S1369-5266(17)30175-9 pmid: 29121612 |
| [34] |
Würschum T, Gross-Hardt R, Laux T. APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem[J]. Plant Cell, 2006, 18(2): 295-307.
doi: 10.1105/tpc.105.038398 pmid: 16387832 |
| [35] |
Lee DY, Lee J, Moon S, et al. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem[J]. Plant J, 2007, 49(1): 64-78.
doi: 10.1111/tpj.2007.49.issue-1 URL |
| [1] |
Xie W, Ding CQ, Hu HT, et al. Molecular events of rice AP2/ERF transcription factors[J]. Int J Mol Sci, 2022, 23(19): 12013.
doi: 10.3390/ijms231912013 URL |
| [2] |
Sharoni AM, Nuruzzaman M, Satoh K, et al. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice[J]. Plant Cell Physiol, 2011, 52(2): 344-360.
doi: 10.1093/pcp/pcq196 pmid: 21169347 |
| [3] |
Allen MD, Yamasaki K, Ohme-Takagi M, et al. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA[J]. EMBO J, 1998, 17(18): 5484-5496.
doi: 10.1093/emboj/17.18.5484 pmid: 9736626 |
| [1] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [2] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [3] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [4] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| [5] | CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics [J]. Biotechnology Bulletin, 2024, 40(3): 135-145. |
| [6] | YANG Wei-cheng, SUN Yan, YANG Qian, WANG Zhuang-lin, MA Ju-hua, XUE Jin-ai, LI Run-zhi. Genome-wide Identification of the FAX family in Gossypium hirsutum and Functional Analysis of GhFAX1 [J]. Biotechnology Bulletin, 2024, 40(3): 155-169. |
| [7] | YANG Wei-jie, YANG Zhou-lin, ZHU Hao-dong, WEI Yu, LIU Jun, LIU Xun. Study on the Properties and Functions of LchAD Protein, a Key Module of Lichenysin Synthase [J]. Biotechnology Bulletin, 2024, 40(3): 322-332. |
| [8] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [9] | GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening [J]. Biotechnology Bulletin, 2024, 40(2): 160-171. |
| [10] | REN Yan-jing, ZHANG Lu-gang, ZHAO Meng-liang, LI Jiang, SHAO Deng-kui. cDNA Yeast Library Construction of Chinese Cabbage Seeds and Screening and Analysis of BrTTG1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(2): 223-232. |
| [11] | XIE Hong, ZHOU Li-ying, LI Shu-wen, WANG Meng-di, AI Ye, CHAO Yue-hui. Structural and Functional Analysis of MtCIM Gene in Medicago truncatula [J]. Biotechnology Bulletin, 2024, 40(1): 262-269. |
| [12] | TANG Wei-lin, KANG Qin, WANG Xia, SHEN Ming-yang, SUN Xin-jiang, WANG Ke, HOU Kai, WU Wei, XU Dong-bei. Cloning and Expression Pattern Analysis of Jasmonic Acid Receptor Gene McCOI1a in Mentha canadensis L. [J]. Biotechnology Bulletin, 2024, 40(1): 270-280. |
| [13] | CHU Rui, LI Zhao-xuan, ZHANG Xue-qing, YANG Dong-ya, CAO Hang-hang, ZHANG Xue-yan. Screening and Identification of Antagonistic Bacillus spp. Against Cucumber Fusarium wilt and Its Biocontrol Effect [J]. Biotechnology Bulletin, 2023, 39(8): 262-271. |
| [14] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [15] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||