








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (7): 237-247.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1099
Previous Articles Next Articles
JIANG Tian-wei1( ), MA Pei-jie2, LI Ya-jiao2, CHEN Cai-jun2, LIU Xiao-xia2, WANG Xiao-li2(
), MA Pei-jie2, LI Ya-jiao2, CHEN Cai-jun2, LIU Xiao-xia2, WANG Xiao-li2( )
)
Received:2024-11-11
Online:2025-07-26
Published:2025-07-22
Contact:
WANG Xiao-li
E-mail:JiangTianwei_GIP@163.com;WangXiaoli_GIP@163.com
JIANG Tian-wei, MA Pei-jie, LI Ya-jiao, CHEN Cai-jun, LIU Xiao-xia, WANG Xiao-li. Metabolic Response Analysis of Brachypodium distachyon to Photoperiods[J]. Biotechnology Bulletin, 2025, 41(7): 237-247.

Fig. 1 Pie chart of metabolite classification, OPLS-DA score plot, and total score statisticsA: Pie chart showing the classification of 739 metabolites. B: OPLS-DA score plot of the five sample groups showing differences. C: OPLS-DA score plot revealing differences between long-day and short-day samples. D: Statistical plot of the total sum of integrals. In Fig. B and C, the X-axis refers to the first principal component score (PC1), which distinguishes different sample groups. The greater the distance, the larger the difference between the groups. The Y-axis refers to the first orthogonal component score (OC1), which reveals the internal correlations of the groups. The smaller the distance between replicates, the higher the reproducibility. In Fig. B and D, LS0 refers to the samples at ZT0, L12 and L24 refer to the samples subjected to long-day treatment for 12 and 24 h, respectively, while S12 and S24 refers to the samples subjected to short-day treatment for 12 and 24 h, respectively. In Fig. C, LS0 is the control group, LD refers to the combined L12 and L24 groups, and SD refers to the combined S12 and S24 groups. In Fig. D, variance analysis was performed using the Tukey multiple comparisons method. Significant differences are indicated by letters, where different letters denote a statistically significant difference (P<0.05). The same below
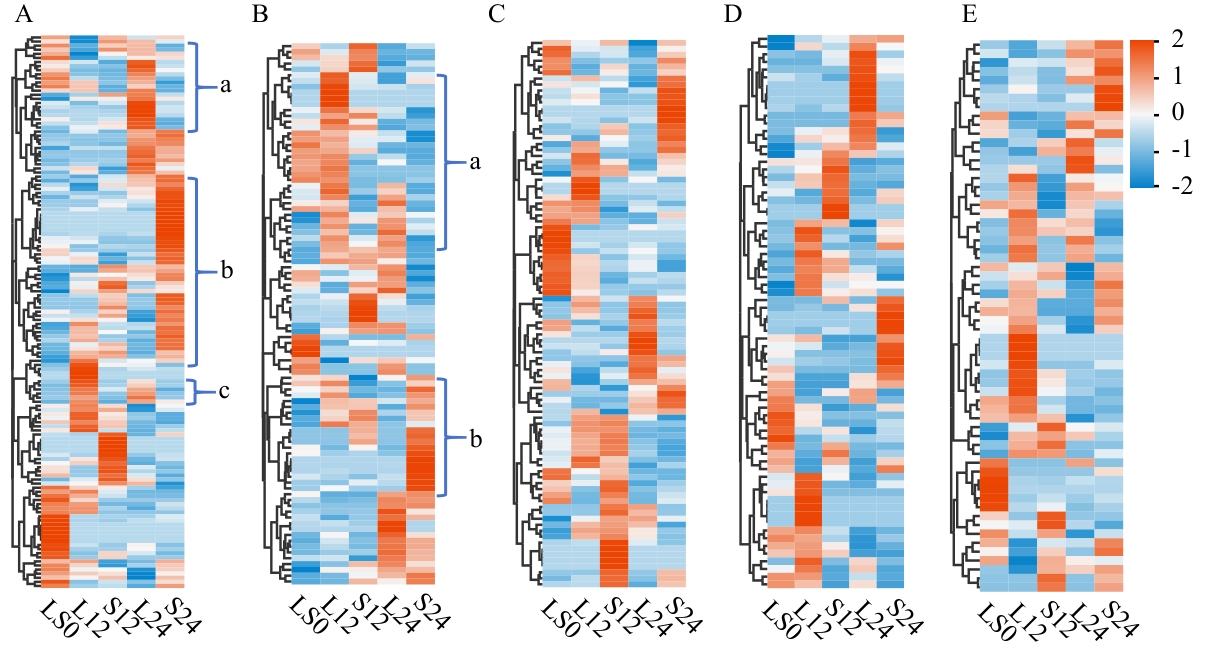
Fig. 2 Clustering heatmap of five classes of metabolitesA: Organic acids and their derivatives. B: Organic oxygen-containing compounds. C: Lipids and lipophilic molecules. D: Phenylpropanoids and polyketides. E: Benzoheterocyclic compounds. Each plot is row-normalized and row-clustered, with red and blue representing upregulation and downregulation, respectively. Lowercase letters a, b, and c indicate regions where metabolites show significant expression differences between long-day and short-day conditions
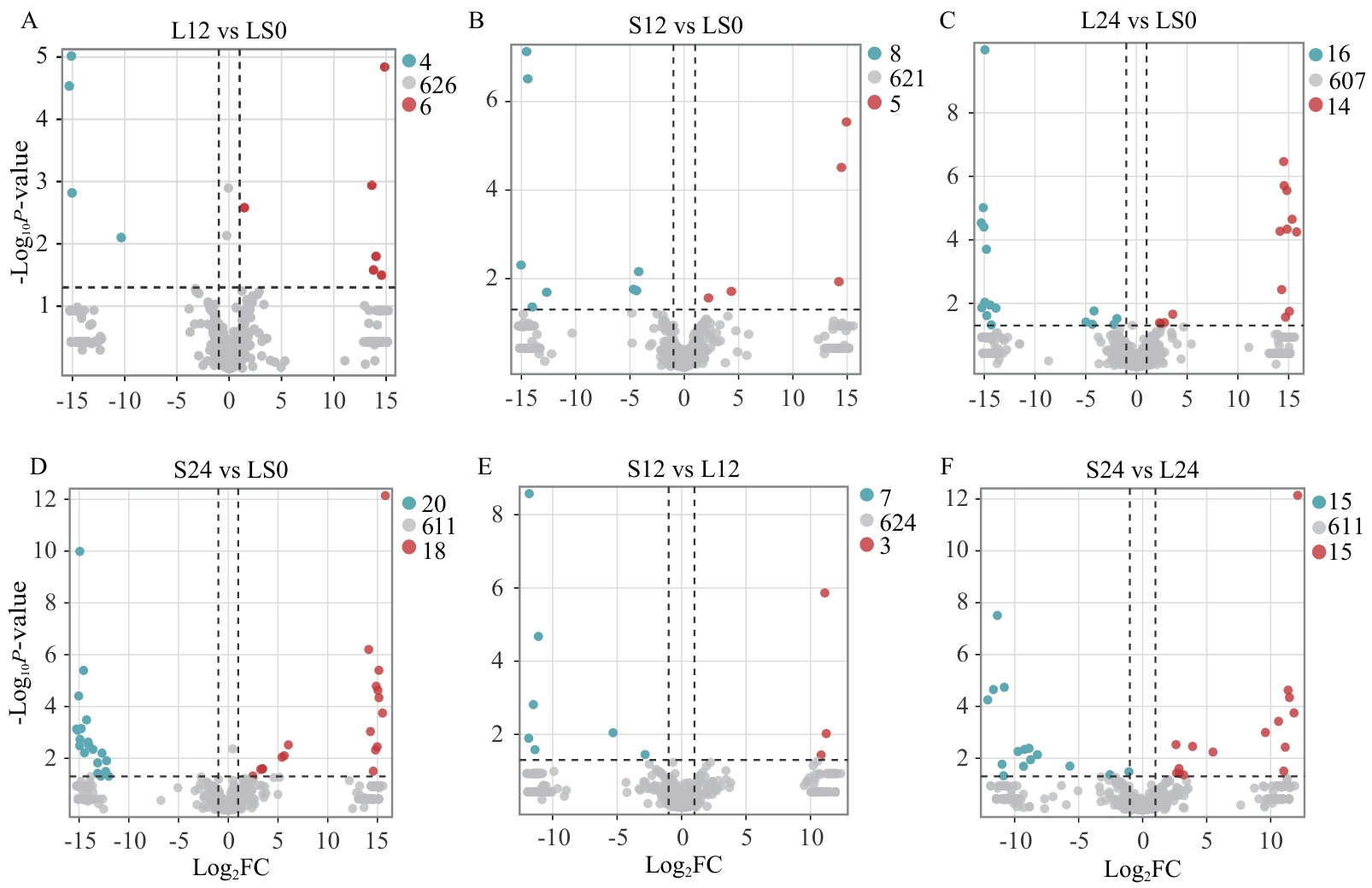
Fig. 3 Volcano plots of differential metabolites for each comparison groupThe x-axis refers to the fold change of each substance in the group comparison, taken as the logarithm base 2. The y-axis indicates the P-value from the t-test, taken as the logarithm base 10. Each dot refers to a metabolite, with red, blue, and gray colors corresponding to significantly up-regulated, significantly down-regulated, and non-significantly changed metabolites, respectively. The dots and numbers in the upper right corner indicate the number of metabolites labeled in different colors
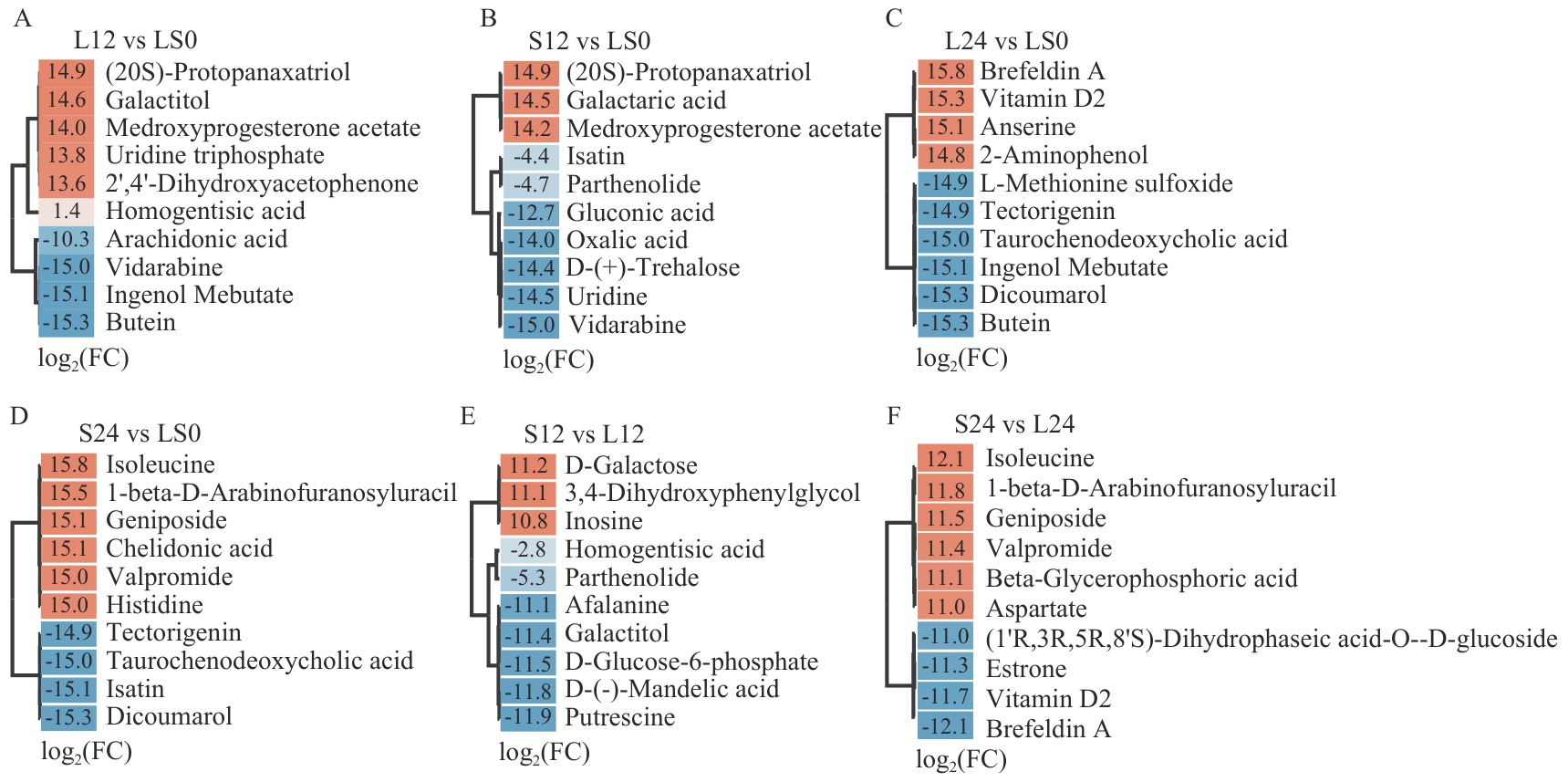
Fig. 4 Top 10 DAMs with the highest fold changes in each comparison groupThe color gradient from light to dark indicates increasing |log2FC| values, with orange for up-regulation and blue for down-regulation. Labels show the |log2FC| values
组合 Group | 代谢物名称 Metabolite | VIP | Log2 倍变比 Log2(FC) | P值 P-value | 上调或下调 Up or down |
|---|---|---|---|---|---|
| S12 vs L12 | 腐胺 Putrescine | 2.08 | 11.86 | 1.28E-02 | DOWN |
| D-(-)-扁桃酸 D-(-)-mandelic acid | 2.30 | 11.82 | 2.65E-09 | DOWN | |
| D-葡萄糖-6-磷酸 D-glucose-6-phosphate | 2.23 | 11.50 | 1.54E-03 | DOWN | |
| 半乳糖醇 Galactitol | 1.98 | 11.37 | 2.62E-02 | DOWN | |
| D-半乳糖 D-galactose | 2.12 | 11.22 | 9.51E-03 | UP | |
| 3,4-二羟基苯乙二醇3,4-dihydroxyphenylglycol | 2.30 | 11.11 | 1.37E-06 | UP | |
| α-丙氨酸 Afalanine | 2.30 | 11.10 | 2.10E-05 | DOWN | |
| 次黄嘌呤核苷 Inosine | 1.94 | 10.83 | 3.65E-02 | UP | |
| 小白菊内酯 Parthenolide | 2.12 | 5.31 | 9.01E-03 | DOWN | |
| 尿黑酸 Homogentisic acid | 1.93 | 2.83 | 3.56E-02 | DOWN | |
| S24 vs L24 | 异亮氨酸 Isoleucine | 2.12 | 12.11 | 7.21E-13 | UP |
| 不枯芽菌素 A Brefeldin A | 2.11 | 12.10 | 5.61E-05 | DOWN | |
| 1-β-D-阿拉伯呋喃糖基尿嘧啶 1-beta-D-arabinofuranosyluracil | 2.10 | 11.82 | 1.79E-04 | UP | |
| 维生素 D2 Vitamin D2 | 2.11 | 11.66 | 2.24E-05 | DOWN | |
| 京尼平苷 Geniposide | 2.11 | 11.47 | 4.54E-05 | UP | |
| 丙戊酰胺 Valpromide | 2.11 | 11.36 | 2.37E-05 | UP | |
| 雌酮 Estrone | 2.12 | 11.35 | 3.09E-08 | DOWN | |
| β-甘油磷酸 Beta-Glycerophosphoric acid | 2.01 | 11.13 | 3.74E-03 | UP | |
| 天冬氨酸 Aspartate | 1.80 | 11.02 | 3.14E-02 | UP | |
| (1'R,3R,5R,8'S)-二氢相酸-O-β-D-葡萄糖苷 (1'R,3R,5R,8'S)-dihydrophaseic acid-O-D-glucoside | 1.89 | 10.99 | 1.71E-02 | DOWN |
Table 1 Top 10 DAM with significant differences between SD and LD conditions at ZT12 and ZT24 in Brachypodium distachyon
组合 Group | 代谢物名称 Metabolite | VIP | Log2 倍变比 Log2(FC) | P值 P-value | 上调或下调 Up or down |
|---|---|---|---|---|---|
| S12 vs L12 | 腐胺 Putrescine | 2.08 | 11.86 | 1.28E-02 | DOWN |
| D-(-)-扁桃酸 D-(-)-mandelic acid | 2.30 | 11.82 | 2.65E-09 | DOWN | |
| D-葡萄糖-6-磷酸 D-glucose-6-phosphate | 2.23 | 11.50 | 1.54E-03 | DOWN | |
| 半乳糖醇 Galactitol | 1.98 | 11.37 | 2.62E-02 | DOWN | |
| D-半乳糖 D-galactose | 2.12 | 11.22 | 9.51E-03 | UP | |
| 3,4-二羟基苯乙二醇3,4-dihydroxyphenylglycol | 2.30 | 11.11 | 1.37E-06 | UP | |
| α-丙氨酸 Afalanine | 2.30 | 11.10 | 2.10E-05 | DOWN | |
| 次黄嘌呤核苷 Inosine | 1.94 | 10.83 | 3.65E-02 | UP | |
| 小白菊内酯 Parthenolide | 2.12 | 5.31 | 9.01E-03 | DOWN | |
| 尿黑酸 Homogentisic acid | 1.93 | 2.83 | 3.56E-02 | DOWN | |
| S24 vs L24 | 异亮氨酸 Isoleucine | 2.12 | 12.11 | 7.21E-13 | UP |
| 不枯芽菌素 A Brefeldin A | 2.11 | 12.10 | 5.61E-05 | DOWN | |
| 1-β-D-阿拉伯呋喃糖基尿嘧啶 1-beta-D-arabinofuranosyluracil | 2.10 | 11.82 | 1.79E-04 | UP | |
| 维生素 D2 Vitamin D2 | 2.11 | 11.66 | 2.24E-05 | DOWN | |
| 京尼平苷 Geniposide | 2.11 | 11.47 | 4.54E-05 | UP | |
| 丙戊酰胺 Valpromide | 2.11 | 11.36 | 2.37E-05 | UP | |
| 雌酮 Estrone | 2.12 | 11.35 | 3.09E-08 | DOWN | |
| β-甘油磷酸 Beta-Glycerophosphoric acid | 2.01 | 11.13 | 3.74E-03 | UP | |
| 天冬氨酸 Aspartate | 1.80 | 11.02 | 3.14E-02 | UP | |
| (1'R,3R,5R,8'S)-二氢相酸-O-β-D-葡萄糖苷 (1'R,3R,5R,8'S)-dihydrophaseic acid-O-D-glucoside | 1.89 | 10.99 | 1.71E-02 | DOWN |
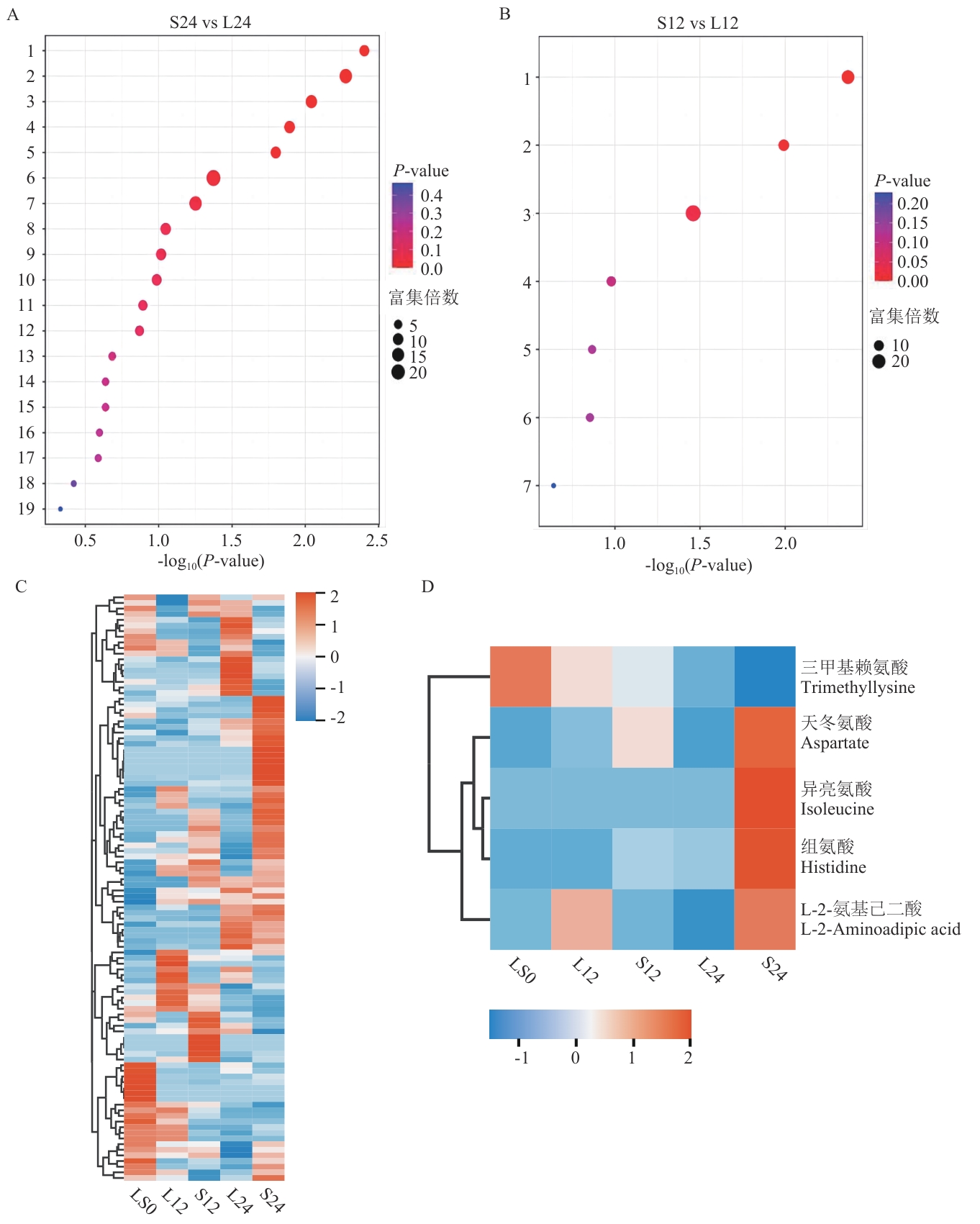
Fig. 5 Enrichment bubble plot of KEGG pathways and heatmap of amino acid-related metabolitesA and B: KEGG pathway enrichment bubble plots for S24 vs L24 and L12 vs S12 groups, respectively. C: Quantitative clustering heatmap of all amino acid metabolites. D: Quantitative clustering heatmap of differential amino acid metabolites. In the bubble plots, larger bubbles indicate a higher number of differentially enriched metabolites in that pathway. In Fig. A, 1: aminoacyl-tRNA biosynthesis; 2: histidine metabolism; 3: β-alanine metabolism; 4: lysine degradation; 5: alanine, aspartate, and glutamate metabolism; 6: phosphoester and phosphorylation metabolism; 7: biosynthesis of valine, leucine, and isoleucine; 8: α-linolenic acid metabolism; 9: arginine biosynthesis; 10: nicotinate and nicotinamide metabolism; 11: pantothenate and CoA biosynthesis; 12: citric acid cycle (TCA cycle); 13: oxalate and dicarboxylate metabolism; 14: glycerophospholipid metabolism; 15: unsaturated fatty acid biosynthesis; 16: degradation of valine, leucine, and isoleucine; 17: tryptophan metabolism; 18: purine metabolism; 19: steroid hormone biosynthesis. In Fig. B, 1: galactose metabolism; 2: tyrosine metabolism; 3: biosynthesis of ubiquinone and other terpenoid-quinones; 4: glutathione metabolism; 5: amino sugar and nucleotide sugar metabolism; 6: arginine and proline metabolism; 7: purine metabolism. Fig. C and D: The heatmaps are row-normalized and clustered, with red indicating upregulation and blue indicating downregulation

Fig. 6 Amino acid-related KEGG metabolic pathway diagramRed boxes indicate pathways, blue shows up-regulated metabolites in S24. Dashed lines indicate indirect superior-subordinate relationships of metabolites, and the solid lines indicate direct relationships. A heat map is drawn for all detected metabolites, red for upregulation, blue for downregulation
| [1] | Osnato M, Cota I, Nebhnani P, et al. Photoperiod control of plant growth: flowering time genes beyond flowering [J]. Front Plant Sci, 2022, 12: 805635. |
| [2] | Mohamed H, Tawfik M, Hussein E, et al. Morphogenic responses of two potato cultivars explants to sucrose, photoperiods and growth regulators [J]. Alfarama J Basic Appl Sci, 2024. |
| [3] | Wafa A, Fekry W, Hassan A, et al. In vitro microtuberization of some potato (Solanum tuberosum L.) cultivars as response to media constituents and photoperiod [J]. Journal of Productivity and Development, 2024, 29(2): 81-98. |
| [4] | Al-Madhagi I. The habit of strawberry flowering is the key for runner propagation, where the photoperiod is the main environmental factor-A review [J]. Adv Hort Sci, 2024, 37(4): 433-449. |
| [5] | Guo L, Plunkert M, Luo X, et al. Developmental regulation of stolon and rhizome [J]. Curr Opin Plant Biol, 2021, 59: 101970. |
| [6] | Lee HG, Jeong YY, Lee H, et al. Arabidopsis HISTONE DEACETYLASE 9 stimulates hypocotyl cell elongation by repressing GIGANTEA expression under short day photoperiod [J]. Front Plant Sci, 2022, 13: 950378. |
| [7] | Xu Y, Koroma AA, Weise SE, et al. Daylength variation affects growth, photosynthesis, leaf metabolism, partitioning, and metabolic fluxes [J]. Plant Physiol, 2023, 194(1): 475-490. |
| [8] | Gendron JM, Staiger D. New horizons in plant photoperiodism [J]. Annu Rev Plant Biol, 2023, 74: 481-509. |
| [9] | Hasterok R, Catalan P, Hazen SP, et al. Brachypodium: 20 years as a grass biology model system; the way forward? [J]. Trends Plant Sci, 2022, 27(10): 1002-1016. |
| [10] | Hasterok R, Marasek A, Donnison IS, et al. Alignment of the genomes of Brachypodium distachyonand temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridization [J]. Genetics, 2006, 173(1): 349-362. |
| [11] | Raissig MT, Woods DP. The wild grass Brachypodium distachyon as a developmental model system [J]. Curr Top Dev Biol, 2022, 147: 33-71. |
| [12] | Ludwig E, Sumner J, Berry J, et al. Natural variation in Brachypodium distachyon responses to combined abiotic stresses [J]. Plant J, 2024, 117(6): 1676-1701. |
| [13] | Suzuki N, Mittler R. Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction [J]. Physiol Plant, 2006, 126(1): 45-51. |
| [14] | Shulaev V, Cortes D, Miller G, et al. Metabolomics for plant stress response [J]. Physiol Plant, 2008, 132(2): 199-208. |
| [15] | Jänkänpää HJ, Mishra Y, Schröder WP, et al. Metabolic profiling reveals metabolic shifts in Arabidopsis plants grown under different light conditions [J]. Plant Cell Environ, 2012, 35(10): 1824-1836. |
| [16] | Ntagkas N, de Vos RCH, Woltering EJ, et al. Modulation of the tomato fruit metabolome by LED light [J]. Metabolites, 2020, 10(6): 266. |
| [17] | Zhan WM, Guo GH, Cui LH, et al. Combined transcriptome and metabolome analysis reveals the effects of light quality on maize hybrids [J]. BMC Plant Biol, 2023, 23(1): 41. |
| [18] | Djerrab D, Bertrand B, Breitler JC, et al. Photoperiod-dependent transcriptional modifications in key metabolic pathways in Coffea arabica [J]. Tree Physiol, 2021, 41(2): 302-316. |
| [19] | Tusevski O, Petreska Stanoeva J, Stefova M, et al. Phenolic profile of dark-grown and photoperiod-exposed Hypericum perforatum L. hairy root cultures [J]. Sci World J, 2013, 2013: 602752. |
| [20] | Yang CJ, Chen W, Tang DD, et al. Metabolomic and transcriptomic insights into anthocyanin biosynthesis in 'ziyan' tea plants under varied photoperiod and temperature conditions [J]. Agronomy, 2024, 14(1): 56. |
| [21] | Yu JY, Yang Y, Luo LJ, et al. Photoperiod-dependent nutrient accumulation in rice cultivated in plant factories: a comparative metabolomic analysis [J]. Foods, 2024, 13(10): 1544. |
| [22] | Vendruscolo RG, Fagundes MB, Maroneze MM, et al. Scenedesmus obliquus metabolomics: effect of photoperiods and cell growth phases [J]. Bioprocess Biosyst Eng, 2019, 42(5): 727-739. |
| [23] | Hoffman DE, Jonsson P, Bylesjö M, et al. Changes in diurnal patterns within the Populus transcriptome and metabolome in response to photoperiod variation [J]. Plant Cell Environ, 2010, 33(8): 1298-1313. |
| [24] | Kumar A, Singh N, Joshi R. Deciphering the metabolic signatures of Trigonella microgreens as a function of photoperiod and temperature using targeted compound analysis and non-targeted UHPLC-QTOF-IMS based approach [J]. Food Res Int, 2024, 176: 113834. |
| [25] | Atif MJ, Amin B, Ghani MI, et al. Transcriptomic analysis of Allium sativum uncovers putative genes involved in photoperiodic pathway and hormone signaling under long day and short day conditions [J]. Plant Sci, 2021, 313: 111095. |
| [26] | Azevedo RA, Arruda P, Turner WL, et al. The biosynthesis and metabolism of the aspartate derived amino acids in higher plants [J]. Phytochemistry, 1997, 46(3): 395-419. |
| [27] | Artins A, Martins MCM, Meyer C, et al. Sensing and regulation of C and N metabolism-novel features and mechanisms of the TOR and SnRK1 signaling pathways [J]. Plant J, 2024, 118(5): 1268-1280. |
| [28] | Lam HM, Coschigano KT, Oliveira IC, et al. The molecular-genetics of nitrogen assimilation into amino acids in higher plants [J]. Annu Rev Plant Physiol Plant Mol Biol, 1996, 47: 569-593. |
| [29] | Richards NG, Schuster SM. An alternative mechanism for the nitrogen transfer reaction in asparagine synthetase [J]. FEBS Lett, 1992, 313(2): 98-102. |
| [30] | Lam HM, Peng SS, Coruzzi GM. Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana [J]. Plant Physiol, 1994, 106(4): 1347-1357. |
| [31] | Lei SH, Rossi S, Huang BR. Metabolic and physiological regulation of aspartic acid-mediated enhancement of heat stress tolerance in perennial ryegrass [J]. Plants, 2022, 11(2): 199. |
| [32] | Han M, Zhang C, Suglo P, et al. L-aspartate: an essential metabolite for plant growth and stress acclimation [J]. Molecules, 2021, 26(7): 1887. |
| [33] | Azevedo RA, Lancien M, Lea PJ. The aspartic acid metabolic pathway, an exciting and essential pathway in plants [J]. Amino Acids, 2006, 30(2): 143-162. |
| [34] | Peng C, Uygun S, Shiu SH, et al. The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis [J]. Plant Physiol, 2015, 169(3): 1807-1820. |
| [35] | Kochevenko A, Araújo WL, Maloney GS, et al. Catabolism of branched chain amino acids supports respiration but not volatile synthesis in tomato fruits [J]. Mol Plant, 2012, 5(2): 366-375. |
| [36] | Janeczko A. Estrogens and androgens in plants: the last 20 years of studies [J]. Plants, 2021, 10(12): 2783. |
| [37] | Dembitsky VM. Biological activity and structural diversity of steroids containing aromatic rings, phosphate groups, or halogen atoms [J]. Molecules, 2023, 28(14): 5549. |
| [38] | Khaleel TF, Dillman R, Gretch D. Estradiol distribution during the development and expression of reproductive structures in Populus tremuloides Michx [J]. Sex Plant Reprod, 2003, 16(1): 35-42. |
| [39] | Janeczko A, Filek W, Biesaga-Kościelniak J, et al. The influence of animal sex hormones on the induction of flowering in Arabidopsis thaliana: comparison with the effect of 24-epibrassinolide [J]. Plant Cell Tissue Organ Cult, 2003, 72(2): 147-151. |
| [40] | Janeczko A, Filek W. Stimulation of generative development in partly vernalized winter wheat by animal sex hormones [J]. Acta Physiol Plant, 2002, 24(3): 291-295. |
| [1] | NIU Ruo-yu, GAO Zhan, XIONG Xian-peng, ZHU De, LUO Hao-tian, MA Xue-yuan, HU Guan-jing. Breeding Applications and Prospects of Wild Cotton Germplasm Resources [J]. Biotechnology Bulletin, 2025, 41(4): 21-32. |
| [2] | HE Cai-lin, LU Jing, GUO Hui-hui, LI Xiao-an, WU Qi. Genome-wide Identification and Expression Analysis of the MADS-box Gene Family in Quinoa [J]. Biotechnology Bulletin, 2025, 41(1): 157-172. |
| [3] | HAN Le-le, SONG Wen-di, BIAN Jia-shen, LI Yang, YANG Shuang-sheng, CHEN Zi-yi, LI Xiao-wei. Revealing the Flavonoid Biosynthesis of Soybean GmERD15c under Salt Stress by Combined Analysis of Transcriptome and Metabolome [J]. Biotechnology Bulletin, 2024, 40(10): 243-252. |
| [4] | JIANG Yu-shan, LAN Qian, WANG Fang, JIANG Liang, PEI Cheng-cheng. Characterization of a Quinoa Mutant Affecting Tyrosine Metabolism [J]. Biotechnology Bulletin, 2024, 40(10): 253-261. |
| [5] | NIU De, HE Yue-hui. Molecular Epigenetic Understanding of Seasonal Regulation of Flowering Time in Wheat [J]. Biotechnology Bulletin, 2024, 40(10): 30-40. |
| [6] | HE Shi-yu, ZENG Zhong-da, LI Bo-yan. Application Progress of Spatially Resolved Metabolomics in Disease Diagnosis Research [J]. Biotechnology Bulletin, 2024, 40(1): 145-159. |
| [7] | ZHOU Ai-ting, PENG Rui-qi, WANG Fang, WU Jian-rong, MA Huan-cheng. Analysis of Metabolic Differences of Biocontrol Strain DZY6715 at Different Growth Stages [J]. Biotechnology Bulletin, 2023, 39(9): 225-235. |
| [8] | HAN Hua-rui, YANG Yu-lu, MEN Yi-han, HAN Shang-ling, HAN Yuan-huai, HUO Yi-qiong, HOU Si-yu. SiYABBYs Involved in Rhamnoside Biosynthesis During the Flower Development of Setaria italica, Based on Metabolomics [J]. Biotechnology Bulletin, 2023, 39(6): 189-198. |
| [9] | XING Yuan, SONG Jian, LI Jun-yi, ZHENG Ting-ting, LIU Si-chen, QIAO Zhi-jun. Identification of AP Gene Family and Its Response Analysis to Abiotic Stress in Setaria italica [J]. Biotechnology Bulletin, 2023, 39(11): 238-251. |
| [10] | XU Yang, DING Hong, ZHANG Guan-chu, GUO Qing, ZHANG Zhi-meng, DAI Liang-xiang. Metabolomics Analysis of Germinating Peanut Seed Under Salt Stress [J]. Biotechnology Bulletin, 2023, 39(1): 199-213. |
| [11] | GULJAMAL·Aisa , XING Jun, LI An, ZHANG Rui. Non-targeted Metabolomics Analysis of Benzo(α)pyrene by Microorganisms in Kefir Grains [J]. Biotechnology Bulletin, 2022, 38(5): 123-135. |
| [12] | ZHU Qiu-yu, DUAN Xu-guo. Recombinant Expression and Site-directed Mutagenesis of L-aspartate-α-decarboxylase,and the Establishment of High-throughput Assay Method [J]. Biotechnology Bulletin, 2022, 38(5): 269-278. |
| [13] | FU Ya-li, PENG Wan-li, LIN Shuang-jun, DENG Zi-xin, LIANG Ru-bing. Gene Cloning and Enzymatic Properties of the Short Chain Dehydrogenase SDR-X1 from Pseudomonas citronellolis SJTE-3 [J]. Biotechnology Bulletin, 2022, 38(3): 121-129. |
| [14] | YANG Yu-ping, ZHANG Xia, WANG Chong-chong, WANG Xiao-yan. Study on Urine Metabolomics in Rats of Different Ages [J]. Biotechnology Bulletin, 2022, 38(2): 166-172. |
| [15] | SUN Shu-fang, LUO Yong-li, LI Chun-hui, JIN Min, XU Qian. Determination of Lignin Monomer Crosslinking Structures in Wheat Stems by UPLC-MS/MS [J]. Biotechnology Bulletin, 2022, 38(10): 66-72. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||