








生物技术通报 ›› 2021, Vol. 37 ›› Issue (12): 180-190.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0274
孟晓建1,2,3( ), 于建东2,3, 郑小梅2,3,4(
), 于建东2,3, 郑小梅2,3,4( ), 郑平2,3,4, 李志敏1(
), 郑平2,3,4, 李志敏1( ), 孙际宾2,3,4, 叶勤1
), 孙际宾2,3,4, 叶勤1
收稿日期:2021-03-10
出版日期:2021-12-26
发布日期:2022-01-19
作者简介:孟晓建,男,硕士研究生,研究方向:酶的代谢调控;E-mail: 基金资助:
MENG Xiao-jian1,2,3( ), YU Jian-dong2,3, ZHENG Xiao-mei2,3,4(
), YU Jian-dong2,3, ZHENG Xiao-mei2,3,4( ), ZHENG Ping2,3,4, LI Zhi-min1(
), ZHENG Ping2,3,4, LI Zhi-min1( ), SUN Ji-bin2,3,4, YE Qin1
), SUN Ji-bin2,3,4, YE Qin1
Received:2021-03-10
Published:2021-12-26
Online:2022-01-19
摘要:
黑曲霉是柠檬酸的工业生产菌株,糖酵解中的3个不可逆反应是柠檬酸积累的重要调控节点,但己糖激酶和丙酮酸激酶的代谢调控研究相对较少,本研究旨在加深对己糖激酶和丙酮酸激酶代谢调控的认识。首先,将黑曲霉己糖激酶和丙酮酸激酶进行内源过表达,经GST亲和层析纯化后重组酶的比酶活分别为(4.86±0.14)U/mg和(1.83±0.02)U/mg。通过小分子代谢物对其酶活影响的检测,发现己糖激酶受果糖-6-磷酸、磷酸烯醇式丙酮酸、ADP和ATP的抑制,而丙酮酸激酶被果糖-1,6-二磷酸、苹果酸和富马酸抑制,同时存在底物ADP的前馈激活作用。进一步通过蛋白三维结构建模与蛋白-小分子代谢物对接模拟分析,发现黑曲霉己糖激酶底物结合位点Asn210及其附近的氨基酸是与小分子代谢物互作的关键位点,而丙酮酸激酶位于别构效应结构域的Thr416、Thr417与Trp470则为关键小分子代谢物的重要结合位点。本研究系统鉴定了小分子代谢物对己糖激酶与丙酮酸激酶的代谢调控关系并预测了其别构调控位点,加深了对黑曲霉糖酵解调控机制的认识。
孟晓建, 于建东, 郑小梅, 郑平, 李志敏, 孙际宾, 叶勤. 小分子代谢物对黑曲霉己糖激酶和丙酮酸激酶的酶活调控[J]. 生物技术通报, 2021, 37(12): 180-190.
MENG Xiao-jian, YU Jian-dong, ZHENG Xiao-mei, ZHENG Ping, LI Zhi-min, SUN Ji-bin, YE Qin. Regulations of Small-molecules Metabolites on Hexokinase and Pyruvate Kinase in Aspergillus niger[J]. Biotechnology Bulletin, 2021, 37(12): 180-190.
| Primer name | Primer sequence(5'-3') | Source |
|---|---|---|
| HxkA-F | gcggcggtggctcctctagaATGGTTGG- AATCGGTCCTAAG | This study |
| HxkA-R | ccgtcgcggtcgactctagaTTATAGCA- GGGTCTTCATGTC | This study |
| PkiA-F | gcggcggtggctcctctagaATGGCCGC- CAGCTCTTCCC | This study |
| PkiA-R | ccgtcgcggtcgactctagaTTACTCAGC- CAGGCCAAG | This study |
| GV-F | CGGAGATTCGTCGCCTAATGTC | This study |
| GV-R | CCGTCGGTCGCAATACAATCAC | This study |
表1 本研究所用的菌株、质粒与引物
Table 1 Strains,plasmids and primers used in this study
| Primer name | Primer sequence(5'-3') | Source |
|---|---|---|
| HxkA-F | gcggcggtggctcctctagaATGGTTGG- AATCGGTCCTAAG | This study |
| HxkA-R | ccgtcgcggtcgactctagaTTATAGCA- GGGTCTTCATGTC | This study |
| PkiA-F | gcggcggtggctcctctagaATGGCCGC- CAGCTCTTCCC | This study |
| PkiA-R | ccgtcgcggtcgactctagaTTACTCAGC- CAGGCCAAG | This study |
| GV-F | CGGAGATTCGTCGCCTAATGTC | This study |
| GV-R | CCGTCGGTCGCAATACAATCAC | This study |
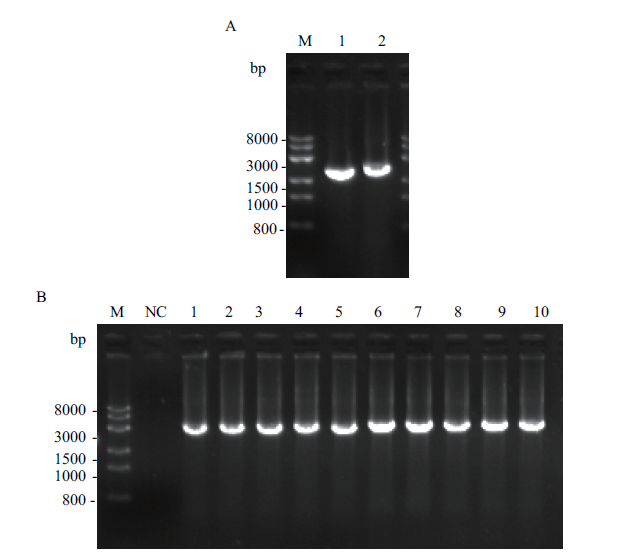
图1 己糖激酶与丙酮酸激酶黑曲霉重组表达菌株构建的PCR扩增电泳图 A:己糖激酶HxkA基因与丙酮酸激酶PkiA基因DNA序列的PCR扩增电泳图,M:Trans 8K DNA marker;1:HxkA基因;2:PkiA基因;B:黑曲霉重组表达菌株AnG1-HxkA与AnG1-PkiA的基因组PCR验证电泳图,M:Trans 8K DNA marker;NC:阴性对照;1-5:AnG1-HxkA各转化子的基因组PCR验证的电泳图,6-10:AnG1-PkiA各转化子的基因组PCR验证的电泳图
Fig. 1 Electrophoretogram of PCR products used for the construction of hexokinase(HxkA)and pyruvate kinase(PkiA)expressed A. niger strains A:PCR products of DNA sequences of hexokinase gene HxkA and pyruvate kinase gene PkiA. M:Trans 8K DNA marker. 1:HxkA gene. 2:PkiA gene. B:Genome PCR verification of recombinant expressing A. niger strain AnG1-HxkA and AnG1-PkiA. M:Trans 8K DNA marker. NC:Negative control. 1-5:The genome PCR verification of the transformants of AnG1-HxkA. 6-10:The genome PCR verification of the transformants of AnG1-PkiA

图2 己糖激酶与丙酮酸激酶GST融合蛋白亲和层析纯化前后的SDS-PAGE电泳图 A:GST-HxkA蛋白亲和层析前后的 SDS-PAGE电泳图,M:蛋白质分子量标准;1:GST-HxkA亲和层析前的SDS-PAGE电泳图;2:GST-HxkA经亲和层析后的SDS-PAGE电泳图;B:GST-PkiA蛋白亲和层析前后的 SDS-PAGE电泳图,M:蛋白质分子量标准;1:GST-PkiA亲和层析前的SDS-PAGE电泳图;2:GST-PkiA经亲和层析后的SDS-PAGE电泳图
Fig.2 SDS-PAGE of the recombinant GST-fused HxkA and PkiA proteins before and after protein purification by affinity chromatography A:SDS-PAGE of GST-HxkA before and after affinity chromatography. M:Protein marker. 1:SDS-PAGE of GST-HxkA before affinity chromatography. 2:SDS-PAGE of recombinant GST-HxkA after affinity chromatography. B:SDS-PAGE of GST-PkiA before and after affinity chromatography. M:Protein marker. 1:SDS-PAGE of GST-PkiA before affinity chromatography. 2:SDS-PAGE of GST-PkiA after affinity chromatography
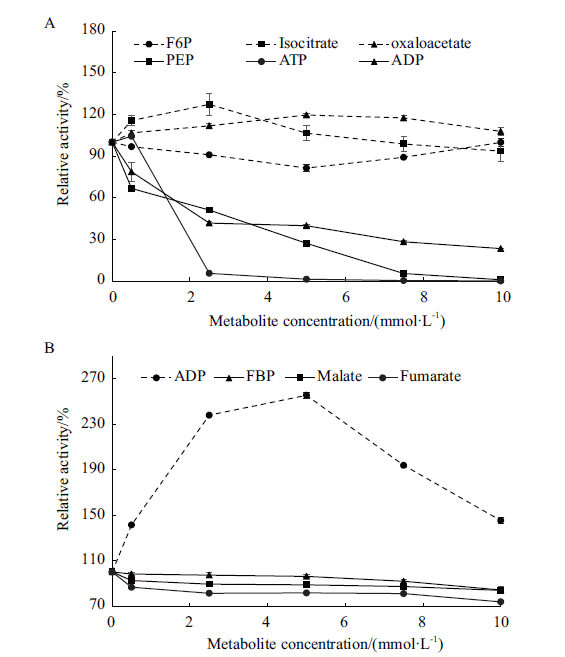
图4 不同浓度的关键小分子代谢物对黑曲霉己糖激酶(A)与丙酮酸激酶酶活(B)影响的检测结果 虚线代表不同浓度的具有激活作用的小分子代谢物存在时酶活变化;实线代表不同浓度的具有抑制作用的小分子代谢物存在时酶活变化
Fig.4 Effects of key metabolites under different concent-rations on the activities of HxkA(A)and PkiA(B)in A. niger The dotted lines refer to changes in enzyme activity in the presence of small molecule metabolites with activation at different concentrations. Solid lines refer to changes in enzyme activity in the presence of small molecule metabolites with inhibition at different concentrations

图5 黑曲霉己糖激酶与丙酮酸激酶的多序列比对结果 A:黑曲霉己糖激酶与不同来源己糖激酶的多序列比对结果,红色五角星为己糖激酶的底物结合位点;B:黑曲霉丙酮酸激酶与不同来源丙酮酸激酶的多序列比对结果,红色五角星为丙酮酸激酶的活性位点
Fig.5 Multiple sequence alignment of HxkA and PkiA from A. niger A:Multiple sequence alignment of HxkA from A. niger with hexokinases from different species,the substrate binding sites of hexokinases are labeled with red stars.B:Multiple sequence alignment of PkiA from A. niger with pyruvate kinases from different species,the catalytic active sites of pyruvate kinases are labeled with red stars

图6 基于同源建模的黑曲霉己糖激酶(A)与丙酮酸激酶(B)三维结构模拟结果 A:蓝色与绿色分别为黑曲霉己糖激酶的2个亚基,红色为黑曲霉己糖激酶的活性位点;B:蓝色、绿色、紫色与黄色分别为黑曲霉丙酮酸激酶的4个亚基,红色为黑曲霉丙酮酸激酶的活性位点
Fig.6 Simulated 3D structure results of HxkA(A)and PkiA(B)from A. niger based on homologous modeling A:Protein structures of 2 sub-units of HxkA in A. niger shown in blue and green;the catalytic active sites of HxkA were labeled in red. B:Protein structures of 4 sub-units of PkiA shown in blue,green,purple and yellow;the catalytic active sites of PkiA are labeled in red

图7 己糖激酶HxkA与关键小分子代谢物的分子对接模拟结果 A:黑曲霉己糖激酶与6-磷酸果糖的分子对接模拟结果;B:己糖激酶与PEP的分子对接模拟结果;C:己糖激酶与ADP的分子对接模拟结果;D:己糖激酶与ATP的分子对接模拟结果。黄色虚线表示氨基酸与关键小分子代谢物形成的氢键,红色数字表示键长
Fig.7 Molecular docking simulation of HxkA with key metabolites A-D:Molecular docking simulation of HxkA with fructose 6-phosphate,PEP,ADP,and ATP,respectively. The hydrogen bonds between the amino acids and key metabolites are represented as yellow dash lines,the hydrogen bond distances are represented in red numbers
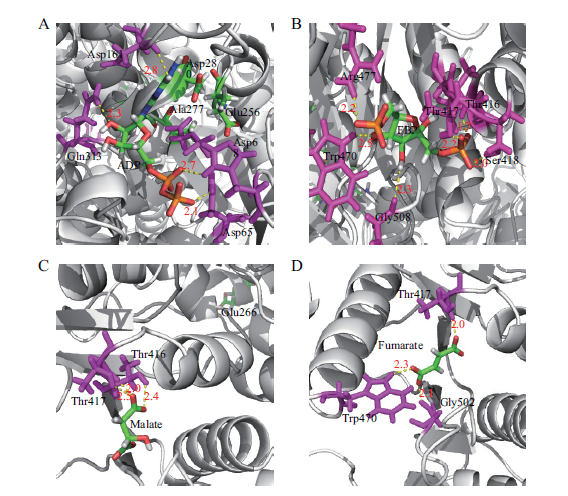
图8 丙酮酸激酶PkiA与关键小分子代谢物的分子对接模拟结果 A:黑曲霉丙酮酸激酶与ADP的分子对接模拟结果;B:丙酮酸激酶与1,6-二磷酸果糖的分子对接模拟结果;C:丙酮酸激酶与苹果酸的分子对接模拟结果;D:丙酮酸激酶与富马酸的分子对接模拟结果。黄色虚线表示氨基酸与关键小分子代谢物形成的氢键,红色数字表示键长。
Fig.8 Molecular docking simulation of PkiA and key metabolites A-D:Molecular docking simulation of PkiA with ADP,fructose 1,6-diphosphate,malic acid,and fumaric acid,respectively. The hydrogen bonds between the amino acids and key metabolites are represented as yellow dash lines;the hydrogen bond distances are represented in red numbers.
| [1] |
Tong Z, Zheng X, Tong Y, et al. Systems metabolic engineering for citric acid production by Aspergillus niger in the post-genomic era[J]. Microb Cell Fact, 2019, 18(1):28.
doi: 10.1186/s12934-019-1064-6 URL |
| [2] |
Anastassiadis S, Morgunov IG, Kamzolova SV, et al. Citric acid production patent review[J]. Recent Patents Biotechnol, 2008, 2(2):107-123.
doi: 10.2174/187220808784619757 URL |
| [3] |
Karaffa L, Kubicek CP. Aspergillus niger citric acid accumulation:do we understand this well working black box?[J]. Appl Microbiol Biotechnol, 2003, 61(3):189-196.
doi: 10.1007/s00253-002-1201-7 URL |
| [4] |
Legisa M, Mattey M. Changes in primary metabolism leading to citric acid overflow in Aspergillus niger[J]. Biotechnol Lett, 2007, 29(2):181-190.
pmid: 17120089 |
| [5] |
Papagianni M. Advances in citric acid fermentation by Aspergillus niger:biochemical aspects, membrane transport and modeling[J]. Biotechnol Adv, 2007, 25(3):244-263.
pmid: 17337335 |
| [6] |
Kubicek-Pranz EM, Mozelt M, Rohr M, et al. Changes in the concentration of fructose 2, 6-bisphosphate in Aspergillus niger during stimulation of acidogenesis by elevated sucrose concentration[J]. Biochim Biophys Acta, 1990, 1033(3):250-255.
pmid: 2156568 |
| [7] |
Mesojednik S, Legisa M. Posttranslational modification of 6-phosphofructo-1-kinase in Aspergillus niger[J]. Appl Environ Microbiol, 2005, 71(3):1425-1432.
doi: 10.1128/AEM.71.3.1425-1432.2005 URL |
| [8] |
Panneman H, Ruijter GJ, van den Broeck HC, et al. Cloning and biochemical characterisation of Aspergillus niger hexokinase——the enzyme is strongly inhibited by physiological concentrations of trehalose 6-phosphate[J]. Eur J Biochem, 1998, 258(1):223-232.
pmid: 9851713 |
| [9] |
Arisan-Atac I, Wolschek MF, Kubicek CP. Trehalose-6-phosphate synthase A affects citrate accumulation by Aspergillus niger under conditions of high glycolytic flux[J]. FEMS Microbiol Lett, 1996, 140(1):77-83.
pmid: 8666204 |
| [10] |
Meixner-Monori B, Kubicek CP, Röhr M. Pyruvate kinase from Aspergillus niger:a regulatory enzyme in glycolysis?[J]. Can J Microbiol, 1984, 30(1):16-22.
pmid: 6713301 |
| [11] |
Zheng X, Zheng P, Zhang K, et al. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger[J]. ACS Synth Biol, 2019, 8(7):1568-1574.
doi: 10.1021/acssynbio.7b00456 URL |
| [12] | Ruijter GJG, Panneman H, Visser J. Overexpression of phosphofructokinase and pyruvate kinase in citric acid-producing Aspergillus niger[J]. Biochim et Biophys Acta, 1997, 1334(2/3):317-326. |
| [13] |
Kuettner EB, Kettner K, Keim A, et al. Crystal structure of hexokinase KlHxk1 of Kluyveromyces lactis:a molecular basis for understanding the control of yeast hexokinase functions via covalent modification and oligomerization[J]. J Biol Chem, 2010, 285(52):41019-41033.
doi: 10.1074/jbc.M110.185850 URL |
| [14] |
Jurica MS, Mesecar A, Heath PJ, et al. The allosteric regulation of pyruvate kinase by fructose-1, 6-bisphosphate[J]. Structure, 1998, 6(2):195-210.
pmid: 9519410 |
| [15] | Land H, Humble MS. YASARA:a tool to obtain structural guidance in biocatalytic investigations[M]. Protein Eng, 2018, 1685:43-67. |
| [16] |
Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server[J]. Nucleic Acids Res, 2014, 42(web server issue):W320-W324.
doi: 10.1093/nar/gku316 URL |
| [17] |
Xu Y, Zhou Y, Cao W, et al. Improved production of malic acid in Aspergillus niger by abolishing citric acid accumulation and enhancing glycolytic flux[J]. ACS Synth Biol, 2020, 9(6):1418-1425.
doi: 10.1021/acssynbio.0c00096 URL |
| [18] |
Mlakar T, Legisa M. Citrate inhibition-resistant form of 6-phosphofructo-1-kinase from Aspergillus niger[J]. Appl Environ Microbiol, 2006, 72(7):4515-4521.
doi: 10.1128/AEM.00539-06 URL |
| [19] |
Yuan M, McNae IW, Chen Y, et al. An allostatic mechanism for M2 pyruvate kinase as an amino-acid sensor[J]. Biochem J, 2018, 475(10):1821-1837.
doi: 10.1042/BCJ20180171 URL |
| [20] |
Susan-Resiga D, Nowak T. Proton donor in yeast pyruvate kinase:chemical and kinetic properties of the active site Thr 298 to Cys mutant[J]. Biochemistry, 2004, 43(48):15230-15245.
pmid: 15568816 |
| [21] |
Bond CJ, Jurica MS, Mesecar A, et al. Determinants of allosteric activation of yeast pyruvate kinase and identification of novel effectors using computational screening[J]. Biochemistry, 2000, 39(50):15333-15343.
pmid: 11112519 |
| [1] | 陈中元, 王玉红, 代为俊, 张艳敏, 叶倩, 刘旭平, 谭文松, 赵亮. 柠檬酸铁铵对悬浮HEK293细胞转染的影响机制探究[J]. 生物技术通报, 2023, 39(9): 311-318. |
| [2] | 成婷, 苑帅, 张晓元, 林良才, 李欣, 张翠英. 酿酒酵母异丁醇合成途径调控的研究进展[J]. 生物技术通报, 2023, 39(7): 80-90. |
| [3] | 段玥彤, 王鹏年, 张春宝, 林春晶. 植物黄烷酮-3-羟化酶基因研究进展[J]. 生物技术通报, 2022, 38(6): 27-33. |
| [4] | 薛鲜丽, 王静然, 毕杭杭, 王德培. 过表达Spt7对黑曲霉生长及抗逆性影响[J]. 生物技术通报, 2022, 38(5): 112-122. |
| [5] | 李毅丹, 单晓辉. 赤霉素代谢调控与绿色革命[J]. 生物技术通报, 2022, 38(2): 195-204. |
| [6] | 田清尹, 岳远征, 申慧敏, 潘多, 杨秀莲, 王良桂. 植物观赏器官中类胡萝卜素代谢调控的研究进展[J]. 生物技术通报, 2022, 38(12): 35-46. |
| [7] | 郭宇飞, 闫荣媚, 张小茹, 曹威, 刘浩. 代谢工程改造黑曲霉生产葡萄糖二酸[J]. 生物技术通报, 2022, 38(11): 227-237. |
| [8] | 曹海鹏, 张书萌, 刁菁, 许拉, 盖春蕾. 一株增强中华绒螯蟹抗病力的固氮红细菌SY5的分离鉴定与表征[J]. 生物技术通报, 2022, 38(11): 277-285. |
| [9] | 袁恺, 何伟, 杨云丽, 朱威宇, 彭超, 安泰, 李丽, 周卫强. 灵芝酸生物合成及代谢调控研究进展[J]. 生物技术通报, 2021, 37(8): 46-54. |
| [10] | 马勤, 雷瑞峰, 迪力热巴·阿不都肉苏力, 穆耶赛尔·奥斯曼, 祖力胡玛尔·肉孜, 安登第. 环境胁迫下内生菌与宿主代谢相互作用研究进展[J]. 生物技术通报, 2021, 37(3): 153-161. |
| [11] | 李红叶, 陈立佼, 刘明丽, 郭天杰, 王道平, 潘映红, 赵明. 黑曲霉单宁酶基因Tan2克隆与表达[J]. 生物技术通报, 2021, 37(3): 44-52. |
| [12] | 高越, 郭晓鹏, 杨阳, 张苗苗, 李文建, 陆栋. 生物丁醇发酵研究进展[J]. 生物技术通报, 2018, 34(8): 27-34. |
| [13] | 陈浩宇, 徐瑞涛, 程志翔, 高强, 张健. H2O2对黑曲霉氧化胁迫机理的研究[J]. 生物技术通报, 2018, 34(4): 201-207. |
| [14] | 徐岩, 韩玉乾, 于放, 刘志文, 王燕燕. 过表达长春花JAR1基因促进文朵灵和长春质碱的生物合成[J]. 生物技术通报, 2017, 33(6): 62-68. |
| [15] | 魏姜勉, 鲁雷震, 焦国宝, 刘家扬, 陆隽鹤. 黑曲霉发酵菌渣对臧红T的吸附研究[J]. 生物技术通报, 2017, 33(10): 191-198. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||