








生物技术通报 ›› 2023, Vol. 39 ›› Issue (1): 187-198.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1033
段敏杰1( ), 李怡斐1, 杨小苗1, 王春萍2, 黄启中1, 黄任中1, 张世才1(
), 李怡斐1, 杨小苗1, 王春萍2, 黄启中1, 黄任中1, 张世才1( )
)
收稿日期:2022-08-23
出版日期:2023-01-26
发布日期:2023-02-02
作者简介:段敏杰,男,硕士,助理研究员,研究方向:加工型辣椒遗传育种;E-mail: 基金资助:
DUAN Min-jie1( ), LI Yi-fei1, YANG Xiao-miao1, WANG Chun-ping2, HUANG Qi-zhong1, HUANG Ren-zhong1, ZHANG Shi-cai1(
), LI Yi-fei1, YANG Xiao-miao1, WANG Chun-ping2, HUANG Qi-zhong1, HUANG Ren-zhong1, ZHANG Shi-cai1( )
)
Received:2022-08-23
Published:2023-01-26
Online:2023-02-02
摘要:
植物DnaJ-Like锌指蛋白(DNAJE)是近年来发现的一类具有重要功能的蛋白,在光合作用、质体发育与运动和植物抗逆及防御反应中发挥重要作用。基于辣椒全基因组数据,对辣椒DNAJE基因家族(CaDNAJE)进行鉴定,利用生物信息学方法对基因和蛋白特征、比较进化、共线性关系及高温胁迫下基因表达模式等进行分析。结果表明,CaDNAJE基因家族有28个成员,不均匀的分布在10条染色体和3个Scaffold上,氨基酸序列长度为99-470 aa,分子量为10.05-52.26 kD,理论等电点(pI)为4.75-9.64,其编码蛋白主要定位在叶绿体和细胞核中;辣椒、番茄和拟南芥的比较进化分析可将其分为GroupⅠ 和GroupⅡ 2个亚族,同一亚族具有类似的蛋白保守motif和基因结构;CaDNAJE基因启动子区包含大量光响应、激素响应和胁迫应答元件;辣椒与拟南芥DNAJE之间存在10对共线性基因,辣椒种内仅有1对;辣椒不同组织转录组分析发现,GroupⅠ 中多数成员在各组织中均有高表达,而GroupⅡ 中除CaDNAJE2、CaDNAJE9等基因存在高表达外,多数基因表达量都很低甚至不表达;高温胁迫下,辣椒叶片过氧化氢含量急速上升后降低,丙二醛含量持续升高,抗氧化物酶活性也有不同程度的提高。同时,高温诱导热激转录因子Hsf和热激蛋白HSP耐热相关基因及CaDNAJE基因高表达,在不同胁迫时期发挥调控作用。推测CaDNAJE基因家族可能参与辣椒响应高温胁迫,提高其耐受力,以降低细胞损伤。
段敏杰, 李怡斐, 杨小苗, 王春萍, 黄启中, 黄任中, 张世才. 辣椒锌指蛋白DnaJ-Like基因家族鉴定及对高温胁迫的表达响应[J]. 生物技术通报, 2023, 39(1): 187-198.
DUAN Min-jie, LI Yi-fei, YANG Xiao-miao, WANG Chun-ping, HUANG Qi-zhong, HUANG Ren-zhong, ZHANG Shi-cai. Identification of Zinc Finger Protein DnaJ-Like Gene Family in Capsicum annuum and Its Expression Analysis Responses to High Temperature Stress[J]. Biotechnology Bulletin, 2023, 39(1): 187-198.
| 基因名称 Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| CaDNAJE2 | TACCCCGAAAAGTGTCCGC | TCGTCGAAAACCACCCTATGAG |
| CaDNAJE3 | TTCTGCTCTATGGGGCTCTTAC | TTTTCCTCCACGGGTTCCT |
| CaDNAJE11 | AACACTCAGCCCTGCTTCCC | GCTGAATGCCACTCCCTTGAC |
| CaDNAJE13 | TGGTGGTGCCGATGAGATT | GCATTCGTCGCAGAGGATAA |
| CaDNAJE15 | CACCAAAGTGAGGAGCATCG | AATCCACCAAGAAATCCAGCA |
| CaDNAJE17 | ATTCCAATCCCAGCCACAA | CAACAGCAGCGATGACCAA |
| CaDNAJE19 | GCTCAGCAAGCGGTAAAT | TGCCATCACCATCCCAGT |
| CaDNAJE21 | CCCTTTCACTTCCAACCCC | TCAGTCCCATTTAGCCCATTTA |
| CaDNAJE23 | CGGTGCCGCTCTGGTGAAATC | CGACAACCCACGTCCCCATCT |
| CaDNAJE24 | TGGGAAGAGTCTATTGATGAACG | ATTGGTAGACACGCTTGGTTTT |
| CaDNAJE27 | ACGCAAAGTAAGAAGAAAGCAGC | CGAGAACAGTATCCAAGCCCAC |
| CaHsfB5 | AAAGTTCCAGAAAGGGTGC | TCCATACATTGAGTGAGGC |
| CaHSP22.0 | CCCCACTCACCATCCCAAAA | CCCCACTGATTCTCAACACCCTA |
| CaHSP16.4 | ATGTCAAAGATGATCAGCTTATTG | GGGTACTCAACACGGGACAC |
| CaUBI3 | GGACCCAAACCCCGATGAT | CACCCCAAGCACAATAAGACA |
表1 实时荧光定量PCR引物
Table 1 Primers used for quantitative real-time PCR
| 基因名称 Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| CaDNAJE2 | TACCCCGAAAAGTGTCCGC | TCGTCGAAAACCACCCTATGAG |
| CaDNAJE3 | TTCTGCTCTATGGGGCTCTTAC | TTTTCCTCCACGGGTTCCT |
| CaDNAJE11 | AACACTCAGCCCTGCTTCCC | GCTGAATGCCACTCCCTTGAC |
| CaDNAJE13 | TGGTGGTGCCGATGAGATT | GCATTCGTCGCAGAGGATAA |
| CaDNAJE15 | CACCAAAGTGAGGAGCATCG | AATCCACCAAGAAATCCAGCA |
| CaDNAJE17 | ATTCCAATCCCAGCCACAA | CAACAGCAGCGATGACCAA |
| CaDNAJE19 | GCTCAGCAAGCGGTAAAT | TGCCATCACCATCCCAGT |
| CaDNAJE21 | CCCTTTCACTTCCAACCCC | TCAGTCCCATTTAGCCCATTTA |
| CaDNAJE23 | CGGTGCCGCTCTGGTGAAATC | CGACAACCCACGTCCCCATCT |
| CaDNAJE24 | TGGGAAGAGTCTATTGATGAACG | ATTGGTAGACACGCTTGGTTTT |
| CaDNAJE27 | ACGCAAAGTAAGAAGAAAGCAGC | CGAGAACAGTATCCAAGCCCAC |
| CaHsfB5 | AAAGTTCCAGAAAGGGTGC | TCCATACATTGAGTGAGGC |
| CaHSP22.0 | CCCCACTCACCATCCCAAAA | CCCCACTGATTCTCAACACCCTA |
| CaHSP16.4 | ATGTCAAAGATGATCAGCTTATTG | GGGTACTCAACACGGGACAC |
| CaUBI3 | GGACCCAAACCCCGATGAT | CACCCCAAGCACAATAAGACA |
| 基序 Motif | 长度 Length/aa | 氨基酸保守基序 Conserved sequence of amino acid |
|---|---|---|
| motif 1 | 20 | CEGCGGSGFVPCSTCNGSGK |
| motif 2 | 33 | EDKIVIYFTSLRGIRKTFEDCNTVRMILESFRV |
| motif 3 | 45 | LQNVLEGKKVSLPRVFIKGRYIGGAEEIKQL- HEEGELKKLLEGJP |
| motif 4 | 15 | CPECNENGLIRCPIC |
| motif 5 | 15 | VDERDVSMDSGFREE |
表2 辣椒DNAJE蛋白氨基酸保守基序
Table 2 Conserved motif of the DNAJE proteins in pepper
| 基序 Motif | 长度 Length/aa | 氨基酸保守基序 Conserved sequence of amino acid |
|---|---|---|
| motif 1 | 20 | CEGCGGSGFVPCSTCNGSGK |
| motif 2 | 33 | EDKIVIYFTSLRGIRKTFEDCNTVRMILESFRV |
| motif 3 | 45 | LQNVLEGKKVSLPRVFIKGRYIGGAEEIKQL- HEEGELKKLLEGJP |
| motif 4 | 15 | CPECNENGLIRCPIC |
| motif 5 | 15 | VDERDVSMDSGFREE |
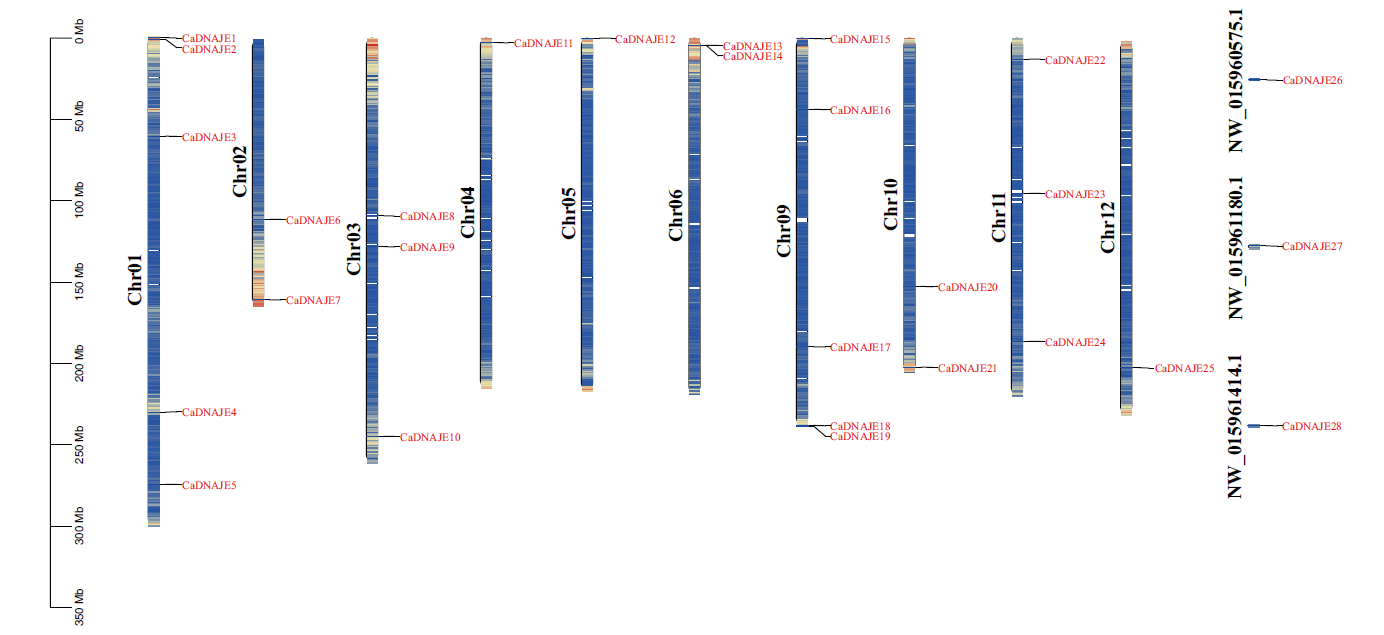
图1 辣椒DNAJE基因家族成员染色体定位 染色体颜色由绿到红表示基因密度由小到大
Fig. 1 Chromosomal location of DNAJE gene family members in pepper(Capsicum annuum) Chromosomal color ranging from green to red indicates the density of gene
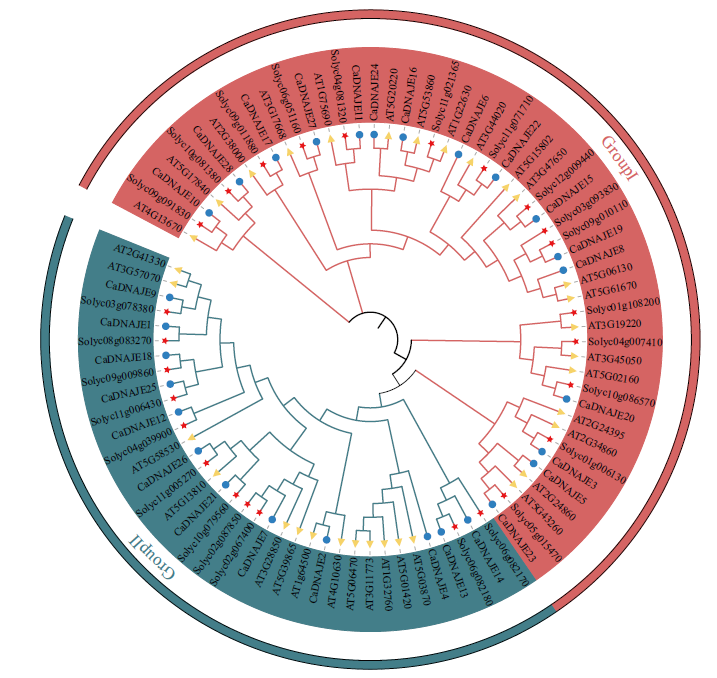
图2 辣椒、拟南芥和番茄DNAJE基因家族成员进化树 图中蓝色实心圆代表辣椒DNAJE蛋白,黄色三角代表拟南芥DNAJE蛋白,红色五角星代表番茄DNAJE蛋白
Fig. 2 Phylogenetic tree of DNAJE gene family members in pepper, Arabidopsis and tomato(Solanum lyc-opersicum) The blue solid dot indicates pepper DNAJE protein, yellow triangle indicates tomato DNAJE protein, and red star indicates Arabidopsis DNAJE protein
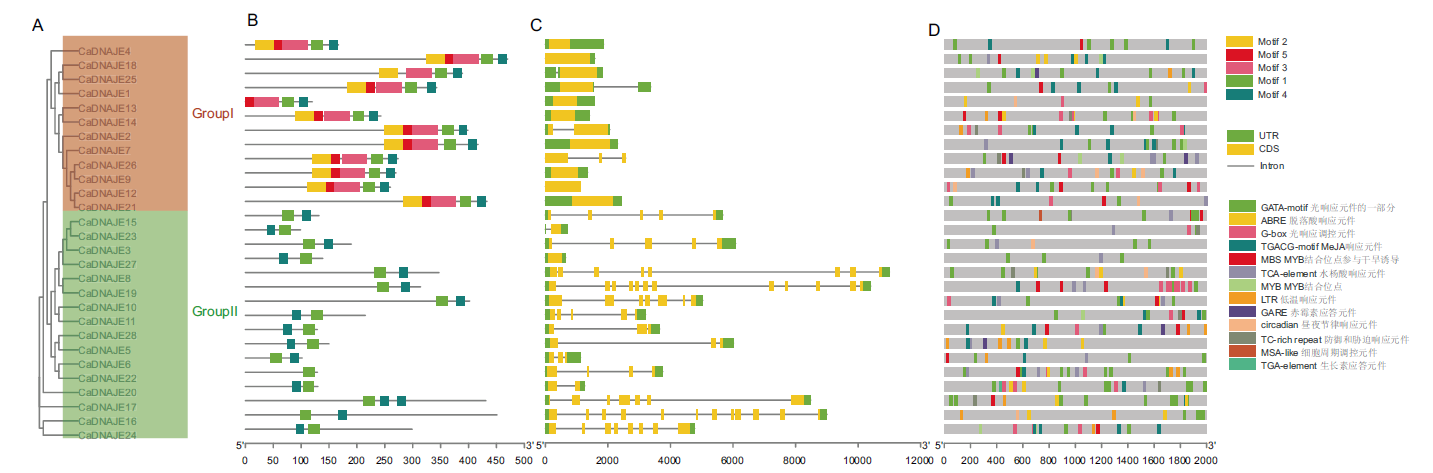
图3 辣椒DNAJE基因家族保守基序、基因结构和顺式作用元件分析 A:CaDNAJE蛋白进化;B:保守基序分布;C:基因结构;D:顺式作用元件分布
Fig. 3 Analysis of conserved motifs, gene structure and cis-acting elements of DNAJE gene family in pepper A: CaDNAJE protein evolution. B: Conserved motif. C: Gene structure. D: cis-acting element distribution
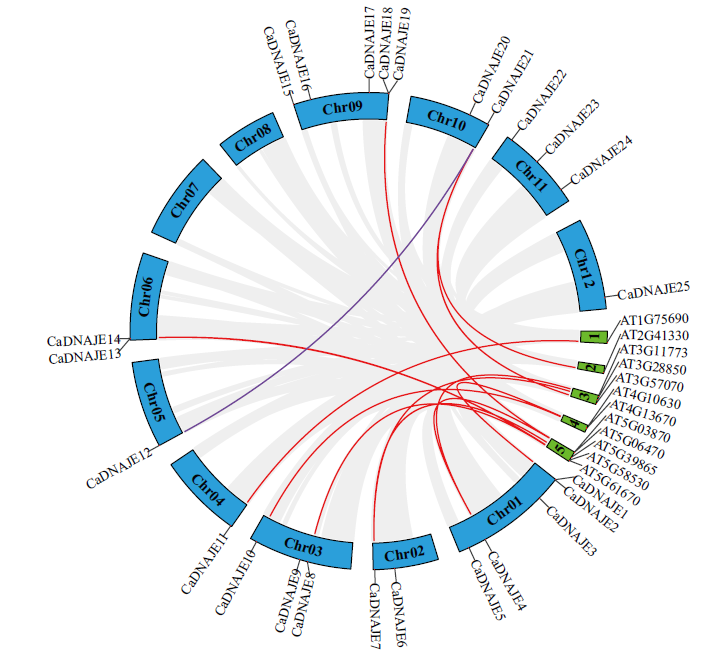
图4 辣椒和拟南芥DNAJE基因家族共线性分析 图中Chr和数字1-5分别表示辣椒和拟南芥的染色体。红线表示种间DNAJE共线性关系,紫线表示种内DNAJE共线性关系,灰线表示种间所有基因成员的共线性关系
Fig. 4 Collinearity analysis of DNAJE gene family between pepper and Arabidopsis Chr and the number 1-5 indicate the chromosomes of pepper and Arabidopsis, respectively. The red lines indicate DNAJE collinearity between interspecific. The purple lines indicate DNAJE collinearity between intraspecific. The Gray lines indicate represent collinearity of all gene membres between interspecific
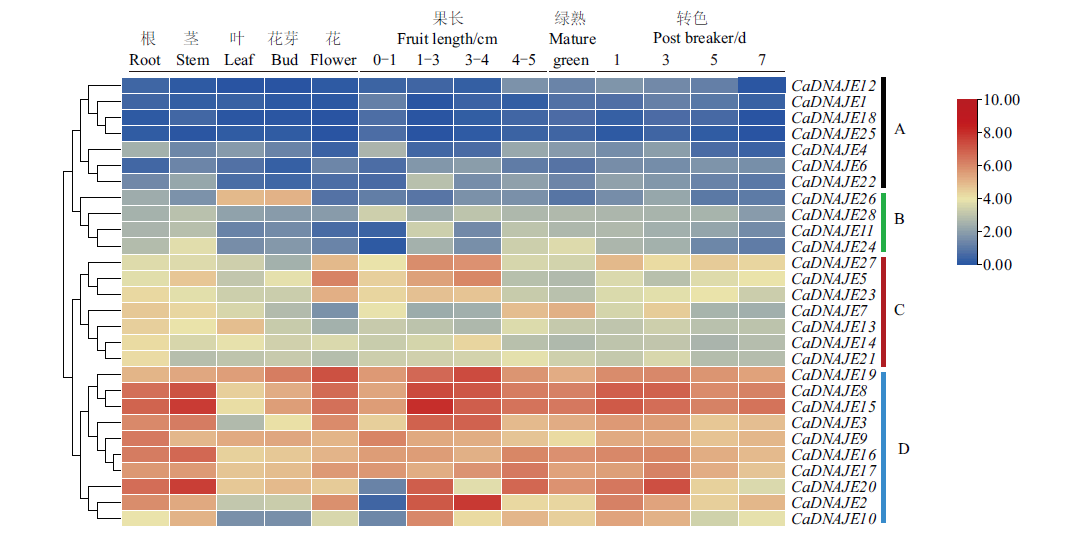
图5 DNAJE基因在辣椒不同组织及果实发育过程中的表达分析 A、B、C、D代表不同表达模式聚类
Fig. 5 Expression profile analysis of pepper DNAJE genes in different tissues and fruit development A, B, C and D indicate the clustering of different expression patterns

图6 辣椒高温胁迫相关基因表达分析 * 表示在0.05水平上差异显著,** 表示在0.01水平上差异显著
Fig. 6 Expression analysis of genes related to high temperature stress in pepper * indicates significant differences at 0.05 level, and ** indicates significant differences at 0.01 level
| [1] |
Georgopoulos CP, Lundquist-Heil A, Yochem J, et al. Identification of the E. coli dnaJ gene product[J]. Mol Gen Genet, 1980, 178(3): 583-588.
doi: 10.1007/BF00337864 URL |
| [2] |
Silver PA, Way JC. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity[J]. Cell, 1993, 74(1): 5-6.
pmid: 8334705 |
| [3] |
Caplan AJ, Cyr DM, Douglas MG. Eukaryotic homologues of Escherichia coli dnaJ: a diverse protein family that functions with hsp70 stress proteins[J]. Mol Biol Cell, 1993, 4(6): 555-563.
pmid: 8374166 |
| [4] |
Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog[J]. J Biol Chem, 1992, 267(29): 20927-20931.
pmid: 1400408 |
| [5] |
Walsh P, Bursac D, Law YC, et al. The J-protein family: modulating protein assembly, disassembly and translocation[J]. EMBO Rep, 2004, 5(6): 567-571.
doi: 10.1038/sj.embor.7400172 pmid: 15170475 |
| [6] |
Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity[J]. Nat Rev Mol Cell Biol, 2010, 11(8): 579-592.
doi: 10.1038/nrm2941 URL |
| [7] |
Ajit Tamadaddi C, Sahi C. J domain independent functions of J proteins[J]. Cell Stress Chaperones, 2016, 21(4): 563-570.
doi: 10.1007/s12192-016-0697-1 URL |
| [8] |
Pulido P, Leister D. Novel DNAJ-related proteins in Arabidopsis thaliana[J]. New Phytol, 2018, 217(2): 480-490.
doi: 10.1111/nph.14827 pmid: 29271039 |
| [9] |
Navrot N, Gelhaye E, Jacquot JP, et al. Identification of a new family of plant proteins loosely related to glutaredoxins with four CxxC motives[J]. Photosynth Res, 2006, 89(2/3): 71-79.
doi: 10.1007/s11120-006-9083-7 URL |
| [10] | 陈斯茗, 卢山. 植物DnaJ-Like锌指蛋白的功能研究[J]. 植物生理学报, 2015, 51(5): 586-592. |
|
Chen SM, Lu S. Recent studies on the DNA J-like zinc finger proteins[J]. Plant Physiol J, 2015, 51(5): 586-592.
doi: 10.1104/pp.51.3.586 URL |
|
| [11] |
Lu S, van Eck J, Zhou XJ, et al. The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation[J]. Plant Cell, 2006, 18(12): 3594-3605.
doi: 10.1105/tpc.106.046417 pmid: 17172359 |
| [12] |
de Saint Germain A, Bonhomme S, Boyer FD, et al. Novel insights into strigolactone distribution and signalling[J]. Curr Opin Plant Biol, 2013, 16(5): 583-589.
doi: 10.1016/j.pbi.2013.06.007 pmid: 23830996 |
| [13] |
Amiya R, Shapira M. ZnJ6 is a thylakoid membrane DnaJ-like chaperone with oxidizing activity in Chlamydomonas reinhardtii[J]. Int J Mol Sci, 2021, 22(3): 1136.
doi: 10.3390/ijms22031136 URL |
| [14] |
Brutnell TP, Sawers RJ, Mant A, et al. BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize[J]. Plant Cell, 1999, 11(5): 849-864.
pmid: 10330470 |
| [15] |
Doron L, Segal N, Gibori H, et al. The BSD2 ortholog in Chlam-ydomonas reinhardtii is a polysome-associated chaperone that co-migrates on sucrose gradients with the rbcL transcript encoding the Rubisco large subunit[J]. Plant J, 2014, 80(2): 345-355.
doi: 10.1111/tpj.12638 URL |
| [16] |
Shimada H, Mochizuki M, Ogura K, et al. Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase[J]. Plant Cell, 2007, 19(10): 3157-3169.
doi: 10.1105/tpc.107.051714 pmid: 17921316 |
| [17] |
Tanz SK, Kilian J, Johnsson C, et al. The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings[J]. Plant J, 2012, 69(5): 743-754.
doi: 10.1111/j.1365-313X.2011.04833.x URL |
| [18] |
Muñoz-Nortes T, Pérez-Pérez JM, Ponce MR, et al. The ANGULA-TA7 gene encodes a DnaJ-like zinc finger-domain protein involved in chloroplast function and leaf development in Arabidopsis[J]. Plant J, 2017, 89(5): 870-884.
doi: 10.1111/tpj.13466 URL |
| [19] |
Albrecht V, Ingenfeld A, Apel K. Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves[J]. Plant Mol Biol, 2008, 66(6): 599-608.
doi: 10.1007/s11103-008-9291-y URL |
| [20] |
Zagari N, Sandoval-Ibañez O, Sandal N, et al. SNOWY COTYLEDON 2 promotes chloroplast development and has a role in leaf variegation in both Lotus japonicus and Arabidopsis thaliana[J]. Mol Plant, 2017, 10(5): 721-734.
doi: 10.1016/j.molp.2017.02.009 URL |
| [21] |
Fristedt R, Williams-Carrier R, Merchant SS, et al. A thylakoid membrane protein harboring a DnaJ-type zinc finger domain is required for photosystem I accumulation in plants[J]. J Biol Chem, 2014, 289(44): 30657-30667.
doi: 10.1074/jbc.M114.587758 pmid: 25228689 |
| [22] |
Lu Y, Hall DA, Last RL. A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana[J]. Plant Cell, 2011, 23(5): 1861-1875.
doi: 10.1105/tpc.111.085456 URL |
| [23] |
Whippo CW, Khurana P, Davis PA, et al. THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility[J]. Curr Biol, 2011, 21(1): 59-64.
doi: 10.1016/j.cub.2010.11.059 pmid: 21185188 |
| [24] |
Zhu JK. Abiotic stress signaling and responses in plants[J]. Cell, 2016, 167(2): 313-324.
doi: 10.1016/j.cell.2016.08.029 URL |
| [25] | 邹学校, 马艳青, 戴雄泽, 等. 辣椒在中国的传播与产业发展[J]. 园艺学报, 2020, 47(9): 1715-1726. |
| Zou XX, Ma YQ, Dai XZ, et al. Spread and industry development of pepper in China[J]. Acta Hortic Sin, 2020, 47(9): 1715-1726. | |
| [26] |
Qin C, Yu C, Shen Y, et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization[J]. PNAS, 2014, 111(14): 5135-5140.
doi: 10.1073/pnas.1400975111 URL |
| [27] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [28] |
Kim S, Park M, Yeom SI, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species[J]. Nat Genet, 2014, 46(3): 270-278.
doi: 10.1038/ng.2877 URL |
| [29] | 樊芳菲. 辣椒DnaJ基因家族:鉴定、结构特征及表达分析[D]. 广州: 华南农业大学, 2017. |
| Fan FF. The DnaJ gene family in pepper(Capsicum annuum L.): comprehensive identification, characterization and expression profiles[D]. Guangzhou: South China Agricultural University, 2017. | |
| [30] |
Wang J, Yang GB, Chen Y, et al. Genome-wide characterization and anthocyanin-related expression analysis of the B-BOX gene family in Capsicum annuum L[J]. Front Genet, 2022, 13: 847328.
doi: 10.3389/fgene.2022.847328 URL |
| [31] |
Arce-Rodríguez ML, Martínez O, Ochoa-Alejo N. Genome-wide identification and analysis of the MYB transcription factor gene family in chili pepper(Capsicum spp.)[J]. Int J Mol Sci, 2021, 22(5): 2229.
doi: 10.3390/ijms22052229 URL |
| [32] |
Jin JH, Wang M, Zhang HX, et al. Genome-wide identification of the AP2/ERF transcription factor family in pepper(Capsicum annuum L.)[J]. Genome, 2018, 61(9): 663-674.
doi: 10.1139/gen-2018-0036 URL |
| [33] |
Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40[J]. Cell Stress Chaperones, 2003, 8(4): 309-316.
doi: 10.1379/1466-1268(2003)008<0309:MFROHF>2.0.CO;2 URL |
| [34] |
Szabo A, Korszun R, Hartl FU, et al. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates[J]. EMBO J, 1996, 15(2): 408-417.
pmid: 8617216 |
| [35] |
Osorio CE. The role of orange gene in carotenoid accumulation: manipulating chromoplasts toward a colored future[J]. Front Plant Sci, 2019, 10: 1235.
doi: 10.3389/fpls.2019.01235 URL |
| [36] |
Zhou XJ, Welsch R, Yang Y, et al. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis[J]. Proc Natl Acad Sci USA, 2015, 112(11): 3558-3563.
doi: 10.1073/pnas.1420831112 URL |
| [37] |
Sun TH, Zhou F, Huang XQ, et al. ORANGE represses chloroplast biogenesis in etiolated Arabidopsis cotyledons via interaction with TCP14[J]. Plant Cell, 2019, 31(12): 2996-3014.
doi: 10.1105/tpc.18.00290 URL |
| [38] |
Jin HQ, Xing MG, Cai CM, et al. B-box proteins in Arachis duranensis: genome-wide characterization and expression profiles analysis[J]. Agronomy, 2019, 10(1): 23.
doi: 10.3390/agronomy10010023 URL |
| [39] |
Hartings S, Paradies S, Karnuth B, et al. The DnaJ-like zinc-finger protein HCF222 is required for thylakoid membrane biogenesis in plants[J]. Plant Physiol, 2017, 174(3): 1807-1824.
doi: 10.1104/pp.17.00401 pmid: 28572458 |
| [40] |
Ham BK, Park JM, Lee SB, et al. Tobacco Tsip1, a DnaJ-type Zn finger protein, is recruited to and potentiates Tsi1-mediated transcriptional activation[J]. Plant Cell, 2006, 18(8): 2005-2020.
doi: 10.1105/tpc.106.043158 URL |
| [41] |
Berman J, Zorrilla-López U, Medina V, et al. The Arabidopsis ORANGE(AtOR)gene promotes carotenoid accumulation in transgenic corn hybrids derived from parental lines with limited carotenoid pools[J]. Plant Cell Rep, 2017, 36(6): 933-945.
doi: 10.1007/s00299-017-2126-z pmid: 28314904 |
| [42] |
Yuan H, Owsiany K, Sheeja TE, et al. A single amino acid substitution in an ORANGE protein promotes carotenoid overaccumulation in Arabidopsis[J]. Plant Physiol, 2015, 169(1): 421-431.
doi: 10.1104/pp.15.00971 pmid: 26224804 |
| [43] |
Zhong LL, Zhou W, Wang HJ, et al. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress[J]. Plant Cell, 2013, 25(8): 2925-2943.
doi: 10.1105/tpc.113.111229 URL |
| [44] | Jin HL, Luo LJ, Wang JF, Wang HB. A novel thylakoid protein, HIGH LIGHT SENSITIVITY1, interacts with LQY1 and involves in photoprotection of photosystemⅡ in Arabidopsis thaliana[C]// 2013 National Congress of Plant Biology, Nanjing, 2013: 248. |
| [45] |
Jacob P, Hirt H, Bendahmane A. The heat-shock protein/chaperone network and multiple stress resistance[J]. Plant Biotechnol J, 2017, 15(4): 405-414.
doi: 10.1111/pbi.12659 pmid: 27860233 |
| [46] |
胡慧芳, 王子雨, 潘潇潇, 等. 辣椒转录因子CaNAC083对高温胁迫的响应[J/OL]. 西北农林科技大学学报: 自然科学版, 2023. DOI:10.13207/j.cnki.jnwafu.2023.03.013.
doi: 10.13207/j.cnki.jnwafu.2023.03.013 |
|
Hu HF, Wang ZY, Pan XX, et al. Response of pepper transcription factor CaNAC083 to high temperature stres[J/OL]. J Northwest A & F Univ Nat Sci Ed, 2023. DOI:10.13207/j.cnki.jnwafu.2023.03.013.
doi: 10.13207/j.cnki.jnwafu.2023.03.013 |
|
| [47] |
Sun JT, Cheng GX, Huang LJ, et al. Modified expression of a heat shock protein gene, CaHSP22.0, results in high sensitivity to heat and salt stress in pepper(Capsicum annuum L.)[J]. Sci Hortic, 2019, 249: 364-373.
doi: 10.1016/j.scienta.2019.02.008 URL |
| [48] |
Huang LJ, Cheng GX, Khan A, et al. CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance[J]. Protoplasma, 2019, 256(1): 39-51.
doi: 10.1007/s00709-018-1280-7 URL |
| [49] | 张驰, 刘丹丹, 刘建中. 植物J蛋白的生物学功能及其作用机制[J]. 浙江大学学报: 农业与生命科学版, 2018, 44(3): 275-282. |
| Zhang C, Liu DD, Liu JZ. Biological functions and action mechanisms of J-domain proteins in plants[J]. J Zhejiang Univ Agric Life Sci, 2018, 44(3): 275-282. | |
| [50] |
Lee KW, Rahman MA, Kim KY, et al. Overexpression of the alfalfa DnaJ-like protein(MsDJLP)gene enhances tolerance to chilling and heat stresses in transgenic tobacco plants[J]. Turk J Biol, 2018, 42(1): 12-22.
doi: 10.3906/biy-1705-30 pmid: 30814866 |
| [51] |
So HA, Chung E, Lee JH. Molecular characterization of soybean GmDjp1 encoding a type III J-protein induced by abiotic stress[J]. Genes Genom, 2013, 35(2): 247-256.
doi: 10.1007/s13258-013-0078-4 URL |
| [1] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [2] | 赵志祥, 王殿东, 周亚林, 王培, 严婉荣, 严蓓, 罗路云, 张卓. 枯草芽孢杆菌Ya-1对辣椒枯萎病的防治及其对根际真菌群落的影响[J]. 生物技术通报, 2023, 39(9): 213-224. |
| [3] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [4] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [5] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [6] | 叶云芳, 田清尹, 施婷婷, 王亮, 岳远征, 杨秀莲, 王良桂. 植物中β-紫罗兰酮生物合成及调控研究进展[J]. 生物技术通报, 2023, 39(8): 91-105. |
| [7] | 张蓓, 任福森, 赵洋, 郭志伟, 孙强, 刘贺娟, 甄俊琦, 王童童, 程相杰. 辣椒响应热胁迫机制的研究进展[J]. 生物技术通报, 2023, 39(7): 37-47. |
| [8] | 李英, 岳祥华. DNA甲基化在解析毛竹自然变异中的应用[J]. 生物技术通报, 2023, 39(7): 48-55. |
| [9] | 成婷, 苑帅, 张晓元, 林良才, 李欣, 张翠英. 酿酒酵母异丁醇合成途径调控的研究进展[J]. 生物技术通报, 2023, 39(7): 80-90. |
| [10] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [11] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [12] | 李敬蕊, 王育博, 解紫薇, 李畅, 吴晓蕾, 宫彬彬, 高洪波. 甜瓜PIN基因家族的鉴定及高温胁迫表达分析[J]. 生物技术通报, 2023, 39(5): 192-204. |
| [13] | 刘奎, 李兴芬, 杨沛欣, 仲昭晨, 曹一博, 张凌云. 青杄转录共激活因子PwMBF1c的功能研究与验证[J]. 生物技术通报, 2023, 39(5): 205-216. |
| [14] | 史建磊, 宰文珊, 苏世闻, 付存念, 熊自立. 番茄青枯病抗性相关miRNA的鉴定与表达分析[J]. 生物技术通报, 2023, 39(5): 233-242. |
| [15] | 周定定, 李辉虎, 汤兴涌, 余发新, 孔丹宇, 刘毅. 甘草酸和甘草苷生物合成与调控的研究进展[J]. 生物技术通报, 2023, 39(5): 44-53. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||