








生物技术通报 ›› 2024, Vol. 40 ›› Issue (12): 193-207.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0503
收稿日期:2024-05-28
出版日期:2024-12-26
发布日期:2025-01-15
通讯作者:
红雨,女,博士,教授,研究方向:修复生态学; E-mail: hongyu@imnu.edu.cn作者简介:殷子薇,女,硕士研究生,研究方向:修复生态学; E-mail: 965078331@qq.com
基金资助:Received:2024-05-28
Published:2024-12-26
Online:2025-01-15
摘要:
【目的】对玫瑰红球菌NB1菌株进行耐盐、促生特性研究,分析其全基因组信息,并挖掘NB1菌株的耐盐促生基因。【方法】利用形态学观察和16S rRNA基因序列分析对NB1菌株进行鉴定。利用固氮菌改良阿须贝氏培养基、Pikovaskaia's培养基、DF液体培养基和ADF液体培养基对NB1菌株的固氮、溶磷以及产ACC脱氨酶能力进行鉴定。将NB1菌株分别接种至盐浓度为0%、5%、10%、15%的NB固体培养基上,培养48 h后确定菌株的可耐受浓度。将经NB1菌株处理和未处理的玉米种子分别接种至1/2 MS培养基上,连续培养15 d后测定其株高、根长、鲜重与根重。以盆栽试验的形式,在同一盐浓度胁迫下,分别测量施加NB1菌液和未处理的玉米幼苗的生长指标。利用Illumina二代测序和PacBio三代测序对NB1菌株进行全基因分析。【结果】NB1菌株鉴定为玫瑰红球菌Rhodococcus rhodochrous,能产生ACC脱氨酶,具有固氮、溶磷等能力,可耐受5%的盐浓度,无土培养条件下,经NB1菌株处理后的玉米幼苗其株高、根长、鲜重和根重均显著增加,盆栽种植后,施加NB1菌液的玉米幼苗株高、鲜重显著高于CK。NB1菌株共编码基因5 259个,编码基因总长度5 230 674 bp,平均GC含量为68.30%。在NR、Pfam、COG、Swiss-Prot、GO、KEGG数据库分别注释到基因5 235、4 379、4 195、3 758、3 263个、2 449个。NB1菌株中有178个基因编码的蛋白质结构属于CAZy家族,内含几丁质酶、蔗糖酶等酶的基因。同时,预测NB1菌株中有14个次级代谢产物基因簇、457个毒力因子、324个耐药基因,且从NB1基因组内发现具有耐盐、促生特性的四氢嘧啶、四环素类抗生素等相关基因。【结论】玫瑰红球菌NB1具有耐盐促生特性,对玉米有一定的促生效果,为植物耐盐促生菌剂提供了新的菌种资源。
殷子薇, 红雨. 玫瑰红球菌NB1对玉米的耐盐促生效应及其全基因组研究[J]. 生物技术通报, 2024, 40(12): 193-207.
YIN Zi-wei, HONG Yu. Study on the Effect of Rhodococcus rhodochrous NB1 on the Tolerance to Salt and Growth-promoting of Maize and Its Whole Genome[J]. Biotechnology Bulletin, 2024, 40(12): 193-207.
| 处理Treatment | 株高Plant height/cm | 根长Root length/cm | 鲜重Fresh weight/g | 根重Root weight/g | |
|---|---|---|---|---|---|
| 无土培养Soil-free culture | CK | 7.21±1.33 | 6.21±1.25 | 1.50±0.13 | 0.40±0.07 |
| NB1 | 10.45±1.12** | 11.76±1.13** | 1.72±0.16* | 0.52±0.07* | |
| 盆栽实验Pot experiment | CK | 24.97±0.96 | 15.98±2.38 | 4.33±0.59 | 2.14±0.32 |
| NB1 | 34.38±2.38** | 19.13±3.38 | 5.29±0.70* | 2.27±0.26 | |
表1 NB1菌液对玉米幼苗生长的影响
Table 1 Effect of NB1 broth on the growth of maize seedlings
| 处理Treatment | 株高Plant height/cm | 根长Root length/cm | 鲜重Fresh weight/g | 根重Root weight/g | |
|---|---|---|---|---|---|
| 无土培养Soil-free culture | CK | 7.21±1.33 | 6.21±1.25 | 1.50±0.13 | 0.40±0.07 |
| NB1 | 10.45±1.12** | 11.76±1.13** | 1.72±0.16* | 0.52±0.07* | |
| 盆栽实验Pot experiment | CK | 24.97±0.96 | 15.98±2.38 | 4.33±0.59 | 2.14±0.32 |
| NB1 | 34.38±2.38** | 19.13±3.38 | 5.29±0.70* | 2.27±0.26 | |

图5 NB1菌株基因组图 圈图的最外面一圈为基因组大小的标识;第二圈和第三圈为正链、负链上的CDS,不同的颜色表示CDS不同的COG的功能分类;第四圈为rRNA和tRNA;第五圈为GC含量,向外的红色部分表示该区域GC含量高于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大,向内的蓝色部分表示该区域GC含量低于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大;最内一圈为GC-Skew值,具体算法为G-C/G+C,可以辅助判断前导链和后滞链,一般前导链GC skew>0,后滞链GC skew<0,也可以辅助判断复制起点(累计偏移最小值)和终点(累计偏移最大值),尤其对环状基因组最为重要
Fig. 5 Genome map of strain NB1 The outermost circle of the circle diagram is the genome size logo. The second and third turns are the CDS on the positive and negative strand. Different colors indicate the functional classification of different COG in CDS. The fourth circle is for the rRNA and the tRNA. The fifth circle shows the GC content. The red portion outward indicates a higher GC content in this region than the genome-wide average GC content. A higher peak indicates a larger difference from the mean GC content. The inward blue part indicates that the GC content in this region is lower than the genome-wide average GC content. A higher peak indicates a greater difference from the mean GC content. The innermost lap is the GC-Skew value, the specific algorithm is G-C/G + C, may assist in the judgment of the leading and lagging strand. The general leading strand, GC skew> 0, the lagging strand GC skew <0, you may also help in judging the replication origin(cumulative offset minimum)and the endpoint(cumulative offset maximum), especially most important for the circular genome
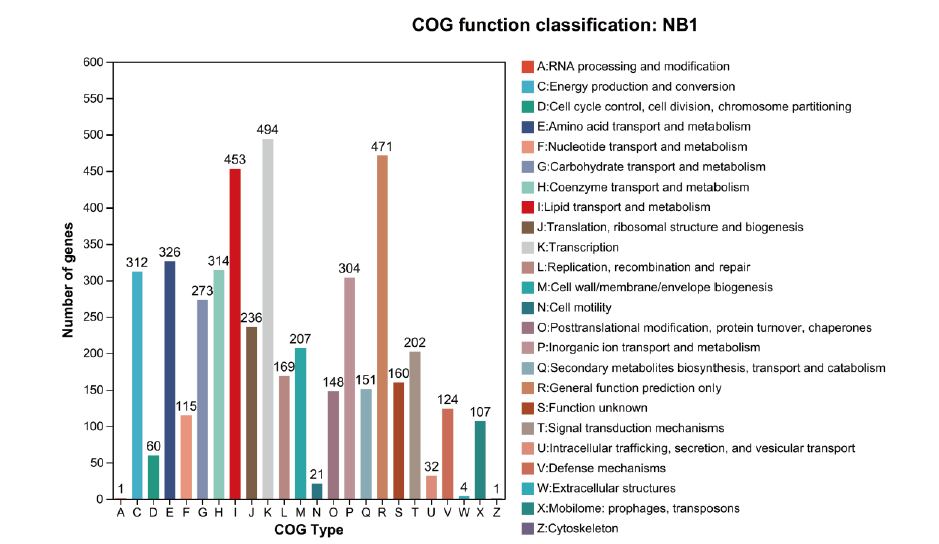
图7 NB1菌株基因组COG数据库比对分析结果 横坐标代表不同COG类型,纵坐标代表基因个数。具体每种COG类型的功能描述请见右侧图例
Fig. 7 Results of COG database alignment analysis of strain NB1 genome The abscissa indicates the different COG types, and the ordinate indicates the number of genes. Specific functional description of each COG type is provided in the legend on the right
| 基因编号Cluster ID | 位置Location | 类型Type | 登录号MIBiG accession | 相似基因簇Similar gene cluster | 相似度Similarity/% | 基因数量Gene No. |
|---|---|---|---|---|---|---|
| Cluster1 | Plasmid B | Butyrolactone | - | - | - | 8 |
| Cluster1 | Chromosome | Terpene | BGC0001456 | Isorenieratene | 25 | 17 |
| Cluster2 | T1PKS | - | - | - | 29 | |
| Cluster3 | Betalactone | - | - | - | 30 | |
| Cluster4 | NAPAA | - | - | - | 30 | |
| Cluster5 | NRPS | BGC0000371 | Heterobactin A / Heterobactin S2 | 36 | 40 | |
| Cluster6 | NRPS | - | - | - | 37 | |
| Cluster7 | Ectoine | BGC0000853 | Ectoine | 75 | 8 | |
| Cluster8 | Terpene | BGC0000269 | SF2575 | 6 | 16 | |
| Cluster9 | NRPS | BGC0000368 | Streptobactin | 11 | 43 | |
| Cluster10 | NRPS | BGC0000893 | Chloramphenicol | 17 | 41 | |
| Cluster11 | Terpene | BGC0000664 | Isorenieratene | 42 | 19 | |
| Cluster12 | NRPS | - | - | - | 39 | |
| Cluster13 | NRPS | - | - | - | 29 |
表2 NB1菌株次级代谢产物合成基因簇
Table 2 Secondary metabolite synthesis gene cluster of strain NB1
| 基因编号Cluster ID | 位置Location | 类型Type | 登录号MIBiG accession | 相似基因簇Similar gene cluster | 相似度Similarity/% | 基因数量Gene No. |
|---|---|---|---|---|---|---|
| Cluster1 | Plasmid B | Butyrolactone | - | - | - | 8 |
| Cluster1 | Chromosome | Terpene | BGC0001456 | Isorenieratene | 25 | 17 |
| Cluster2 | T1PKS | - | - | - | 29 | |
| Cluster3 | Betalactone | - | - | - | 30 | |
| Cluster4 | NAPAA | - | - | - | 30 | |
| Cluster5 | NRPS | BGC0000371 | Heterobactin A / Heterobactin S2 | 36 | 40 | |
| Cluster6 | NRPS | - | - | - | 37 | |
| Cluster7 | Ectoine | BGC0000853 | Ectoine | 75 | 8 | |
| Cluster8 | Terpene | BGC0000269 | SF2575 | 6 | 16 | |
| Cluster9 | NRPS | BGC0000368 | Streptobactin | 11 | 43 | |
| Cluster10 | NRPS | BGC0000893 | Chloramphenicol | 17 | 41 | |
| Cluster11 | Terpene | BGC0000664 | Isorenieratene | 42 | 19 | |
| Cluster12 | NRPS | - | - | - | 39 | |
| Cluster13 | NRPS | - | - | - | 29 |
| 药物类别Drug class | 基因数量Gene No. | 药物类别Drug class | 基因数量Gene No. | |
|---|---|---|---|---|
| Disinfecting agents and antiseptics | 57 | Polyamine antibiotic | 6 | |
| Tetracycline antibiotic | 51 | Elfamycin antibiotic | 5 | |
| Macrolide antibiotic | 42 | Pleuromutilin antibiotic | 4 | |
| Fluoroquinolone antibiotic | 31 | Streptogramin antibiotic | 4 | |
| Peptide antibiotic | 30 | Fusidane antibiotic | 3 | |
| Penam | 24 | Sulfonamide antibiotic | 3 | |
| Cephalosporin | 22 | Thioamide antibiotic | 3 | |
| Rifamycin antibiotic | 22 | Diaminopyrimidine antibiotic | 3 | |
| Phenicol antibiotic | 21 | Penem | 3 | |
| Aminoglycoside antibiotic | 20 | Salicylic acid antibiotic | 3 | |
| Phosphonic acid antibiotic | 18 | Nitroimidazole antibiotic | 3 | |
| Glycopeptide antibiotic | 16 | Mupirocin-like antibiotic | 3 | |
| Aminocoumarin antibiotic | 15 | Nucleoside antibiotic | 2 | |
| Isoniazid-like antibiotic | 13 | Sulfone antibiotic | 2 | |
| Carbapenem | 13 | Pyrazine antibiotic | 2 | |
| Oxazolidinone antibiotic | 13 | Antibacterial free fatty acids | 2 | |
| Cephamycin | 10 | Nitrofuran antibiotic | 1 | |
| Glycylcycline | 9 | Streptogramin B antibiotic | 1 | |
| Monobactam | 7 | Streptogramin A antibiotic | 1 | |
| Lincosamide antibiotic | 7 | Nybomycin-like antibiotic | 1 |
表3 NB1菌株耐药基因预测分析统计
Table 3 Statistics on prediction analysis of resistance genes in strain NB1
| 药物类别Drug class | 基因数量Gene No. | 药物类别Drug class | 基因数量Gene No. | |
|---|---|---|---|---|
| Disinfecting agents and antiseptics | 57 | Polyamine antibiotic | 6 | |
| Tetracycline antibiotic | 51 | Elfamycin antibiotic | 5 | |
| Macrolide antibiotic | 42 | Pleuromutilin antibiotic | 4 | |
| Fluoroquinolone antibiotic | 31 | Streptogramin antibiotic | 4 | |
| Peptide antibiotic | 30 | Fusidane antibiotic | 3 | |
| Penam | 24 | Sulfonamide antibiotic | 3 | |
| Cephalosporin | 22 | Thioamide antibiotic | 3 | |
| Rifamycin antibiotic | 22 | Diaminopyrimidine antibiotic | 3 | |
| Phenicol antibiotic | 21 | Penem | 3 | |
| Aminoglycoside antibiotic | 20 | Salicylic acid antibiotic | 3 | |
| Phosphonic acid antibiotic | 18 | Nitroimidazole antibiotic | 3 | |
| Glycopeptide antibiotic | 16 | Mupirocin-like antibiotic | 3 | |
| Aminocoumarin antibiotic | 15 | Nucleoside antibiotic | 2 | |
| Isoniazid-like antibiotic | 13 | Sulfone antibiotic | 2 | |
| Carbapenem | 13 | Pyrazine antibiotic | 2 | |
| Oxazolidinone antibiotic | 13 | Antibacterial free fatty acids | 2 | |
| Cephamycin | 10 | Nitrofuran antibiotic | 1 | |
| Glycylcycline | 9 | Streptogramin B antibiotic | 1 | |
| Monobactam | 7 | Streptogramin A antibiotic | 1 | |
| Lincosamide antibiotic | 7 | Nybomycin-like antibiotic | 1 |
| [1] | 王振营, 王晓鸣. 我国玉米病虫害发生现状、趋势与防控对策[J]. 植物保护, 2019, 45(01):1-11. |
| Wang ZY, Wang XO. Current status and management strategies for corn pests anddiseases in China[J]. Plant Protection, 2019, 45(1): 1-11. | |
| [2] | 王晓光, 史桂清, 刘春阁, 等. 中国青贮玉米产业现状及发展趋势[J]. 农学学报, 2023, 13(7): 20-24. |
| Wang XG, Shi GQ, Liu CG, et al. Present situation and development trend of silage corn industry in China[J]. Journal of Agriculture, 2023, 13(7): 20-24. | |
| [3] | 沈彦. 盐碱环境胁迫对吸胀萌发期玉米种子SOD活性的影响[J]. 绿色科技, 2022, 24(13): 121-122, 126. |
| Shen Y. Effect of saline alkli environmental stresses on SOD antioxidant enzymes in maize seeds during germination[J]. J Green Sci Technol, 2022, 24(13): 121-122, 126. | |
| [4] |
冯玉倩, 米俊珍, 赵宝平, 等. 秸秆配施微生物菌肥对盐碱地土壤及作物盐分含量的影响[J]. 华北农学报, 2023, 38(6): 101-107.
doi: 10.7668/hbnxb.20193788 |
| Feng YQ, Mi JZ, Zhao BP, et al. Effect of straw combined with microbial fertilizer on salt content of soil and crops in saline alkali land[J]. Acta Agric Boreali Sin, 2023, 38(6): 101-107. | |
| [5] |
王亚文, 史慧芳, 张鹏, 等. 微生物菌肥在设施蔬菜生产中的研究进展[J]. 农学学报, 2021, 11(11): 27-32.
doi: 10.11923/j.issn.2095-4050.casb2021-0063 |
|
Wang YW, Shi HF, Zhang P, et al. Research progress of microbial fertilizer in facility vegetable production[J]. J Agric, 2021, 11(11): 27-32.
doi: 10.11923/j.issn.2095-4050.casb2021-0063 |
|
| [6] | 冯健茹, 俄文慧, 朱秀玲, 等. 一株栖稻假单胞菌NYCS1-5的鉴定及其对玉米的耐盐促生功能[J]. 山东农业大学学报: 自然科学版, 2021, 52(5): 723-730. |
| Feng JR, E WH, Zhu XL, et al. Identification of a strain Pseu-domonas oryzihabitans NYCS1-5 and its growth-promoting function for maize on high saline condition[J]. J Shandong Agric Univ Nat Sci Ed, 2021, 52(5): 723-730. | |
| [7] | 张芬芬, 周晓伦, 贺洋洋. 一株溶磷促生菌的分离、鉴定及其对玉米幼苗生长的影响[J]. 广东农业科学, 2021, 48(5): 76-82. |
| Zhang FF, Zhou XL, He YY. Isolation and identification of a phosphorus-solubilizing growth-promoting bacterium and its effect on the growth of Zea mays L. seedlings[J]. Guangdong Agric Sci, 2021, 48(5): 76-82. | |
| [8] | Sarr M, Seiler J, Sullivan J. Growth and physiology of Senegalia senegal(L.) britton seedlings as influenced by seed origin and salinity and fertility treatments[J]. Forests, 2017, 8(10): 388. |
| [9] | Rashid MI, Shah GA, Iqbal Z, et al. Nanobiochar associated ammonia emission mitigation and toxicity to soil microbial biomass and corn nutrient uptake from farmyard manure[J]. Plants, 2023, 12(9): 1740. |
| [10] | 岳思君, 李海荣, 武珍珍, 等. 硒砂瓜连作地微生物群落变化及一株优势芽孢杆菌的分离鉴定[J]. 中国瓜菜, 2016, 29(12): 19-22, 37. |
| Yue SJ, Li HR, Wu ZZ, et al. Changes of microbial community and isolation and identification of a dominant bacillus strain from the continuous cropping of selenium sand watermelon[J]. China Cucurbits Veg, 2016, 29(12): 19-22, 37. | |
| [11] | 杨杉杉, 李国光, 张胜男, 等. 假单胞菌BP16的分离鉴定及其植物促生性状和效应[J]. 微生物学通报, 2018, 45(10): 2121-2130. |
| Yang SS, Li GG, Zhang SN, et al. Isolation and identification of Pseudomonas sp. BP16 and its plant growth-promoting traits and effects[J]. Microbiol China, 2018, 45(10): 2121-2130. | |
| [12] | Zhang FL, Huang N, Li L, et al. Screening of acetochlor-resistance -promoting bacteria and their growth-promoting effeets on maize seedlings[J/OL]. Journal of Agricultural Resources and Environment, 2024, 41(1): 212. |
| [13] | 姚拓. 高寒地区燕麦根际联合固氮菌研究II固氮菌的溶磷性和分泌植物生长素特性测定[J]. 草业学报, 2004, 13(3): 85-90. |
| Yao T. Associative nitrogen-fixing bacteria in the rhizosphere of Avena sativa in an alpine region II Phosphate-solubilizing power and auxin production[J]. Acta Prataculturae Sin, 2004, 13(3): 85-90. | |
| [14] | 杨苗. 具有ACC脱氨酶活性促生细菌的筛选、鉴定及其接种效应研究[D]. 杨凌: 西北农林科技大学, 2018. |
| Yang M. Screening, identification and inoculation effect of growth-promoting bacteria with ACC deaminase activity[D]. Yangling: Northwest A & F University, 2018. | |
| [15] | 吕杰, 周晓馥, 未晓巍, 等. 基因型及基本培养基对玉米成熟胚培养的影响[J]. 安徽农业科学, 2011, 39(29): 17767-17768, 17780. |
| Lv J, Zhou XF, Wei XW, et al. Effects of genotypes and basic medium on culture of maize mature embryos[J]. Journal of Anhui Agri, 2011, 39(29):17767-17768, 17780. | |
| [16] | Luo RB, Liu BH, Xie YL, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler[J]. GigaScience, 2012, 1(1): 18. |
| [17] | Wick RR, Judd LM, Gorrie CL, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads[J]. PLoS Comput Biol, 2017, 13(6): e1005595. |
| [18] |
Delcher AL, Bratke KA, Powers EC, et al. Identifying bacterial genes and endosymbiont DNA with Glimmer[J]. Bioinformatics, 2007, 23(6): 673-679.
doi: 10.1093/bioinformatics/btm009 pmid: 17237039 |
| [19] | Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses[J]. Nucleic Acids Res, 2005, 33(Web Server issue): W451-W454. |
| [20] |
Chan PP, Lowe TM. tRNAscan-SE: searching for tRNA genes in genomic sequences[J]. Methods Mol Biol, 2019, 1962: 1-14.
doi: 10.1007/978-1-4939-9173-0_1 pmid: 31020551 |
| [21] |
Benson G. Tandem repeats finder: a program to analyze DNA sequences[J]. Nucleic Acids Res, 1999, 27(2): 573-580.
doi: 10.1093/nar/27.2.573 pmid: 9862982 |
| [22] |
Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics[J]. Genome Res, 2009, 19(9): 1639-1645.
doi: 10.1101/gr.092759.109 pmid: 19541911 |
| [23] |
Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000[J]. Nucleic Acids Res, 2000, 28(1): 45-48.
doi: 10.1093/nar/28.1.45 pmid: 10592178 |
| [24] | Finn RD, Bateman A, Clements J, et al. Pfam: the protein families database[J]. Nucleic Acids Res, 2014, 42(Database issue): D222-D230. |
| [25] | Jensen LJ, Julien P, Kuhn M, et al. eggNOG: automated construction and annotation of orthologous groups of genes[J]. Nucleic Acids Res, 2008, 36(Database issue): D250-D254. |
| [26] |
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Res, 2000, 28(1): 27-30.
doi: 10.1093/nar/28.1.27 pmid: 10592173 |
| [27] | Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database(CAZy)in 2013[J]. Nucleic Acids Res, 2014, 42(Database issue): D490-D495. |
| [28] | Blin K, Wolf T, Chevrette MG, et al. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification[J]. Nucleic Acids Res, 2017, 45(W1): W36-W41. |
| [29] | Chen LH, Zheng DD, Liu B, et al. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on[J]. Nucleic Acids Res, 2016, 44(D1): D694-D697. |
| [30] | Jia BF, Raphenya AR, Alcock B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database[J]. Nucleic Acids Res, 2017, 45(D1): D566-D573. |
| [31] | 侯冬梅, 张兰, 李春成, 等. 一株耐盐异养硝化-好氧反硝化菌Rhodococcus sp. LS-2的分离鉴定与脱氮性能研究[J]. 南昌航空大学学报:自然科学版, 2023, 37(3): 50-58, 102. |
| Hou DM, Zhang L, Li CC, et al. Isolation, identification and nitrogen removal performance of a halotolerant heterotrophic nitrification-aerobic denitrification bacteria Rhodococcus sp.LS-2[J]. Journal of Nanchang Hangkong University: Natural Sciences, 2023, 37(3): 50-58, 102. | |
| [32] | 卢惠娇. 一株降解PHBA红球菌的筛选、鉴定及其生物活性研究[D]. 南宁: 广西民族大学, 2023. |
| Lu HJ. Screening, identification and biological activity of a Rhodococcus sp. degrading PHBA[D]. Nanning: Guangxi University for Nationalities, 2023. | |
| [33] | 王小松. T.guizhouense NJAU4742几丁质酶Chi8的特性及其水解产物对植物促生效果研究[D]. 南京: 南京农业大学, 2020. |
| Wang XS. Study on the characteristics of chitinase Chi8 from T.guizhouense NJAU4742 and the effect of its hydrolysate on plant growth promotion[D]. Nanjing: Nanjing Agricultural University, 2020. | |
| [34] | 郭嘉, 陈纪香, 于一雷, 等. 黄河三角洲湿地典型盐生植物群落土壤酶活性研究[J]. 湿地科学与管理, 2020, 16(1): 55-59. |
| Guo J, Chen JX, Yu YL, et al. Characterization of soil enzyme activities of six typical halophyte plant communities in the wetland of Yellow River Delta[J]. Wetl Sci Manag, 2020, 16(1): 55-59. | |
| [35] | Kotroczó Z, Veres Z, Fekete I, et al. Soil enzyme activity in response to long-term organic matter manipulation[J]. Soil Biol Biochem, 2014, 70: 237-243. |
| [36] | Xu HX, Chen YT, Huang JZ, et al. Advances in ectoine biosynthesis and biochemical characteristics of key enzymes[J]. Chinese Journal of Biotechuology, 2024, 40(6): 1620-1643. |
| [37] | 张山, 胡萌, 何永志, 等. 四氢嘧啶微生物合成与应用研究进展[J]. 微生物学报, 2021, 61(8): 2250-2263. |
| Zhang S, Hu M, He YZ, et al. Research progress in microbial production and application of ectoines[J]. Acta Microbiol Sin, 2021, 61(8): 2250-2263. | |
| [38] | Yu HS, Bo G, Yan D, et al. Isolation and ectoine-producing characteristics of halophiles from soil of Jilantai Saline Lake[J]. J Northwest A&F Univ, 2019, 47: 115-131. |
| [39] | 翁南海, 王焕宇, 张磊, 等. 一株耐盐反硝化细菌的筛选、鉴定和特性及其产物四氢嘧啶的检测[J]. 微生物学通报, 2023, 50(6): 2335-2348. |
| Weng NH, Wang HY, Zhang L, et al. A salt-tolerant denitrifying bacterial strain: screening,identification, characterization, and detection of its product ectoine[J]. Microbiology China, 2023, 50(6): 2335-2348. | |
| [40] | Ma YH, Huang P, Huang SC, et al. γ-Aminobutyric acid(GABA)and ectoine(ECT)impacts with and without AMF on antioxidants, gas exchange attributes and nutrients of cotton cultivated in salt affected soil[J]. BMC Plant Biol, 2023, 23(1): 476. |
| [41] | Ren HF. In vitro growth inhibition effects of enterobactin-specific antibodies on different gram-negative bacteria[J]. Progress in Microbiology and Immunology, 2021, 49(5): 28. |
| [42] | 李佳珣, 叶雨寒, 胡心怡, 等. 抑制铜绿假单胞菌和金黄色葡萄球菌生物膜形成的乳杆菌筛选及机制初探[J]. 微生物学杂志, 2023, 43(6): 89-96. |
| Li JX, Ye YH, Hu XY, et al. Sereening of biofilm forming lactobacillus inhibiting Pseudomonas aeruginosa and Staphylococcus aureus and initial probe of its mechanism[J]. Journal of Microbiology, 2023, 43(6): 89-96. | |
| [43] | 邓佳琪. 四环素类抗生素对藜麦生长发育及产量的影响[D]. 临汾: 山西师范大学, 2022. |
| Deng JQ. Effects of tetracycline antibiotics on growth and yield of quinoa[D]. Linfen: Shanxi Normal University, 2022. |
| [1] | 何财林, 卢晶, 郭会会, 李小安, 吴琪. 藜麦MADS-box基因家族的全基因组鉴定和表达分析[J]. 生物技术通报, 2025, 41(1): 157-172. |
| [2] | 张婷, 万雨欣, 徐伟慧, 王志刚, 陈文晶, 胡云龙. 一株玉米根际促生菌Leclercia adecarboxylata LN01促生效果研究及其基因组分析[J]. 生物技术通报, 2025, 41(1): 263-275. |
| [3] | 崔原瑗, 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 大麦非特异性磷脂酶C基因家族全基因组鉴定及苗期胁迫表达分析[J]. 生物技术通报, 2024, 40(8): 74-82. |
| [4] | 孙志勇, 杜怀东, 刘阳, 马嘉欣, 于雪然, 马伟, 姚鑫杰, 王敏, 李培富. 水稻籽粒γ-氨基丁酸含量的全基因组关联分析[J]. 生物技术通报, 2024, 40(8): 53-62. |
| [5] | 任晓敏, 云岚, 艾芊, 赵乔. 新麦草异戊烯基转移酶PjIPT基因的功能验证[J]. 生物技术通报, 2024, 40(7): 207-215. |
| [6] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| [7] | 周江鸿, 夏菲, 仲丽, 仇兰芬, 李广, 刘倩, 张国锋, 邵金丽, 李娜, 车少臣. 黄栌枯萎病拮抗细菌CCBC3-3-1的全基因组测序及比较基因组分析[J]. 生物技术通报, 2024, 40(7): 235-246. |
| [8] | 田彤彤, 葛家振, 高鹏程, 李学瑞, 宋国栋, 郑福英, 储岳峰. 绵羊肺炎支原体GH3-3株全基因组测序及生物信息学分析[J]. 生物技术通报, 2024, 40(7): 323-334. |
| [9] | 王秋月, 段鹏亮, 李海笑, 刘宁, 曹志艳, 董金皋. 玉米大斑病菌cDNA文库的构建及转录因子StMR1互作蛋白的筛选[J]. 生物技术通报, 2024, 40(6): 281-289. |
| [10] | 阿丽亚·外力, 陈永坤, 克拉热木·克里木江, 王宝庆, 陈凌娜. 核桃SPL基因家族的系统进化和表达分析[J]. 生物技术通报, 2024, 40(6): 180-189. |
| [11] | 胡锦锦, 李素贞, 马旭辉, 柳小庆, 谢珊珊, 江海洋, 陈茹梅. 玉米花青素生物合成代谢调控[J]. 生物技术通报, 2024, 40(6): 34-44. |
| [12] | 孙亚楠, 王春雪, 王欣, 杜秉海, 刘凯, 汪城墙. 萎缩芽孢杆菌CNY01的生防特性及其对玉米的抗盐促生作用[J]. 生物技术通报, 2024, 40(5): 248-260. |
| [13] | 孔小平, 陈利文, 刘思思, 严湘萍. 胡萝卜抽薹相关性状全基因组关联分析[J]. 生物技术通报, 2024, 40(5): 120-130. |
| [14] | 王佳玮, 李晨, 刘建利, 周世杰, 易嘉敏, 杨谨源, 康鹏. 内生真菌接种方式对青贮玉米幼苗生长的影响[J]. 生物技术通报, 2024, 40(4): 189-202. |
| [15] | 徐伟芳, 李贺宇, 张慧, 何仔昂, 高文恒, 谢紫洋, 王传文, 尹登科. 生防细菌HX0037对栝楼炭疽病的防病能力及其机制[J]. 生物技术通报, 2024, 40(4): 228-241. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
