








生物技术通报 ›› 2022, Vol. 38 ›› Issue (1): 77-85.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0181
收稿日期:2021-02-13
出版日期:2022-01-26
发布日期:2022-02-22
作者简介:吕迪,男,硕士研究生,研究方向:生物化学与分子生物学;E-mail: 基金资助:
LV Di( ), CHEN Ru-mei, ZHOU Xiao-jin(
), CHEN Ru-mei, ZHOU Xiao-jin( )
)
Received:2021-02-13
Published:2022-01-26
Online:2022-02-22
摘要:
JAZ(jasmonate ZIM-domain)蛋白是茉莉酸(jasmonic acid,JA)信号途径中关键的负调控因子,明确JAZ蛋白和MYC2之间的结合关系对整个JA信号通路至关重要。通过qRT-PCR筛选了在籽粒中较特异表达的ZmJAZ4、生殖器官中高表达的ZmJAZ8以及组成型表达的ZmJAZ12,利用玉米叶肉细胞原生质体瞬时表达体系,通过亚细胞定位和双分子荧光互作(bimolecular fluorescence complementation)研究了这些JAZ蛋白的定位以及是否可以与ZmMYC2相互作用。研究结果发现ZmJAZ4、ZmJAZ8和ZmJAZ12均定位于细胞核,同时BiFC结果显示这些JAZ蛋白都可与ZmMYC2在细胞核中相互作用,证实了ZmJAZ家族蛋白可与JA信号通路关键转录因子ZmMYC2结合的功能,提示它们可通过与ZmMYC2的结合影响其转录调控活性。
吕迪, 陈茹梅, 周晓今. ZmJAZ与ZmMYC2的BiFC互作研究[J]. 生物技术通报, 2022, 38(1): 77-85.
LV Di, CHEN Ru-mei, ZHOU Xiao-jin. Interactions Between ZmJAZ and ZmMYC2 Using Bimolecular Fluorescence Complementation Assay[J]. Biotechnology Bulletin, 2022, 38(1): 77-85.
| 基因名称 Gene name | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| ZmActin | ZmActin-qF | ATGTTTCCTGGGATTGCCGAT |
| ZmActin-qR | CCAGTTTCGTCATACTCTCCCTTG | |
| ZmJAZ4 | ZmJAZ4-qF | CGCTCAGGCTCTGAAACAGTGAAACC |
| ZmJAZ4-qR | CGACGACAGACACAGTGCCTAAGAAT | |
| ZmJAZ8 | ZmJAZ8-qF | TTGAGAAACGCCGTGACAGGGT |
| ZmJAZ8-qR | GTCAGCCTTCACCGATGGGACT | |
| ZmJAZ12 | ZmJAZ12-qF | CTGATGCTAAGAAGCCTACTCGC |
| ZmJAZ12-qR | GCGTCTGAAGGAGAAGTTTGGTA |
表1 实时荧光定量PCR所用引物
Table 1 Primers used in qRT-PCR
| 基因名称 Gene name | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| ZmActin | ZmActin-qF | ATGTTTCCTGGGATTGCCGAT |
| ZmActin-qR | CCAGTTTCGTCATACTCTCCCTTG | |
| ZmJAZ4 | ZmJAZ4-qF | CGCTCAGGCTCTGAAACAGTGAAACC |
| ZmJAZ4-qR | CGACGACAGACACAGTGCCTAAGAAT | |
| ZmJAZ8 | ZmJAZ8-qF | TTGAGAAACGCCGTGACAGGGT |
| ZmJAZ8-qR | GTCAGCCTTCACCGATGGGACT | |
| ZmJAZ12 | ZmJAZ12-qF | CTGATGCTAAGAAGCCTACTCGC |
| ZmJAZ12-qR | GCGTCTGAAGGAGAAGTTTGGTA |
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 扩增产物 Product |
|---|---|---|
| ZmMYC2-F | ATGAACCTGTGGACGGAC | ZmMYC2-ORF |
| ZmMYC2-R | TTACCTGCCCATGGCAGACCCG | |
| ZmJAZ4-F | ATGGCCGCCTCCGGGAACAACA | ZmJAZ4-ORF |
| ZmJAZ4-R | TCAGAGCCTGAGCGCGAGCCACGA | |
| ZmJAZ8-F | ATGGATCTGTTGGAGCGGAATATT | ZmJAZ8-ORF |
| ZmJAZ8-R | TTACTGGGCCTTGCCCTCCGCTGT | |
| ZmJAZ12-F | ATGGCGGCGTCCGCGAGGCCCG | ZmJAZ12-ORF |
| ZmJAZ12-R | CTAGCTCAGGTTCAGGTTGGACTT | |
| ZmMYC2-GF | TCATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGACGACAA | ZmMYC2-GFP |
| ZmMYC2-GR | CCTCGCCCTTGCTCACCATTCTAGACCTGCCCATGGCAGACCCGGGCT | |
| ZmJAZ4-GF | ATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAACA | ZmJAZ4-GFP |
| ZmJAZ4-GR | CCTCGCCCTTGCTCACCATTCTAGAGAGCCTGAGCGCGAGCCAC | |
| ZmJAZ8-GF | ATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAATA | ZmJAZ8- GFP |
| ZmJAZ8-GR | CCTCGCCCTTGCTCACCATTCTAGACTGGGCCTTGCCCTCCGCT | |
| ZmJAZ12-GF | ATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-GFP |
| ZmJAZ12-GR | CCTCGCCCTTGCTCACCATCTAGAGCTCAGGTTCAGGTTGGACT | |
| ZmMYC2-NF | ATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGAC | ZmMYC2-YFPN |
| ZmMYC2-NR | CTTTTGCTCCATTCTAGACCTGCCCATGGCAGACCC | |
| ZmMYC2-NF | ATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGAC | ZmMYC2-YFPC |
| ZmMYC2-CR | GTATGGGTACATTCTAGACCTGCCCATGGCAGACCC | |
| ZmJAZ4-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAA | ZmJAZ4-YFPN |
| ZmJAZ4-NR | AACTTTTGCTCCATTCTAGAGAGCCTGAGCGCGAGCCACGAGG | |
| ZmJAZ4-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAA | ZmJAZ4-YFPC |
| ZmJAZ4-CR | CATCGTATGGGTACATTCTAGAGAGCCTGAGCGCGAGCCACGAGG | |
| ZmJAZ8-NF | CATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAAT | ZmJAZ8-YFPN |
| ZmJAZ8-NR | AACTTTTGCTCCATTCTAGACTGGGCCTTGCCCTCCGCTGTCAC | |
| ZmJAZ8-NF | CATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAAT | ZmJAZ8-YFPC |
| ZmJAZ8-CR | CATCGTATGGGTACATTCTAGACTGGGCCTTGCCCTCCGCTGTCAC | |
| ZmJAZ12-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-YFPN |
| ZmJAZ12-NR | AACTTTTGCTCCATTCTAGAGCTCAGGTTCAGGTTGGACTTGAC | |
| ZmJAZ12-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-YFPC |
| ZmJAZ12-CR | CATCGTATGGGTACATTCTAGAGCTCAGGTTCAGGTTGGACTTGAC |
表2 PCR扩增所用引物
Table 2 Primers used for gene cloning
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 扩增产物 Product |
|---|---|---|
| ZmMYC2-F | ATGAACCTGTGGACGGAC | ZmMYC2-ORF |
| ZmMYC2-R | TTACCTGCCCATGGCAGACCCG | |
| ZmJAZ4-F | ATGGCCGCCTCCGGGAACAACA | ZmJAZ4-ORF |
| ZmJAZ4-R | TCAGAGCCTGAGCGCGAGCCACGA | |
| ZmJAZ8-F | ATGGATCTGTTGGAGCGGAATATT | ZmJAZ8-ORF |
| ZmJAZ8-R | TTACTGGGCCTTGCCCTCCGCTGT | |
| ZmJAZ12-F | ATGGCGGCGTCCGCGAGGCCCG | ZmJAZ12-ORF |
| ZmJAZ12-R | CTAGCTCAGGTTCAGGTTGGACTT | |
| ZmMYC2-GF | TCATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGACGACAA | ZmMYC2-GFP |
| ZmMYC2-GR | CCTCGCCCTTGCTCACCATTCTAGACCTGCCCATGGCAGACCCGGGCT | |
| ZmJAZ4-GF | ATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAACA | ZmJAZ4-GFP |
| ZmJAZ4-GR | CCTCGCCCTTGCTCACCATTCTAGAGAGCCTGAGCGCGAGCCAC | |
| ZmJAZ8-GF | ATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAATA | ZmJAZ8- GFP |
| ZmJAZ8-GR | CCTCGCCCTTGCTCACCATTCTAGACTGGGCCTTGCCCTCCGCT | |
| ZmJAZ12-GF | ATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-GFP |
| ZmJAZ12-GR | CCTCGCCCTTGCTCACCATCTAGAGCTCAGGTTCAGGTTGGACT | |
| ZmMYC2-NF | ATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGAC | ZmMYC2-YFPN |
| ZmMYC2-NR | CTTTTGCTCCATTCTAGACCTGCCCATGGCAGACCC | |
| ZmMYC2-NF | ATTTGGAGAGGACCTCGAGATGAACCTGTGGACGGAC | ZmMYC2-YFPC |
| ZmMYC2-CR | GTATGGGTACATTCTAGACCTGCCCATGGCAGACCC | |
| ZmJAZ4-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAA | ZmJAZ4-YFPN |
| ZmJAZ4-NR | AACTTTTGCTCCATTCTAGAGAGCCTGAGCGCGAGCCACGAGG | |
| ZmJAZ4-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCCGCCTCCGGGAACAA | ZmJAZ4-YFPC |
| ZmJAZ4-CR | CATCGTATGGGTACATTCTAGAGAGCCTGAGCGCGAGCCACGAGG | |
| ZmJAZ8-NF | CATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAAT | ZmJAZ8-YFPN |
| ZmJAZ8-NR | AACTTTTGCTCCATTCTAGACTGGGCCTTGCCCTCCGCTGTCAC | |
| ZmJAZ8-NF | CATTTCATTTGGAGAGGACCTCGAGATGGATCTGTTGGAGCGGAAT | ZmJAZ8-YFPC |
| ZmJAZ8-CR | CATCGTATGGGTACATTCTAGACTGGGCCTTGCCCTCCGCTGTCAC | |
| ZmJAZ12-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-YFPN |
| ZmJAZ12-NR | AACTTTTGCTCCATTCTAGAGCTCAGGTTCAGGTTGGACTTGAC | |
| ZmJAZ12-NF | CATTTCATTTGGAGAGGACCTCGAGATGGCGGCGTCCGCGAGGCC | ZmJAZ12-YFPC |
| ZmJAZ12-CR | CATCGTATGGGTACATTCTAGAGCTCAGGTTCAGGTTGGACTTGAC |

图1 ZmJAZ4、ZmJAZ8和ZmJAZ12基因的相对表达量 Root:根;Stem:茎;Leaf:叶片;Tassel:未散粉的雄花;Ear:未授粉的雌穗;En:胚乳;Em:胚;DAP(Days after pollination):授粉后天数;数据以平均值±标准差表示
Fig.1 Relative expressions of ZmJAZ4,ZmJAZ8,and ZmJAZ12 En:Endosperm. Em:Embryo. DAP:Day after pollination. The data are in means ± SD
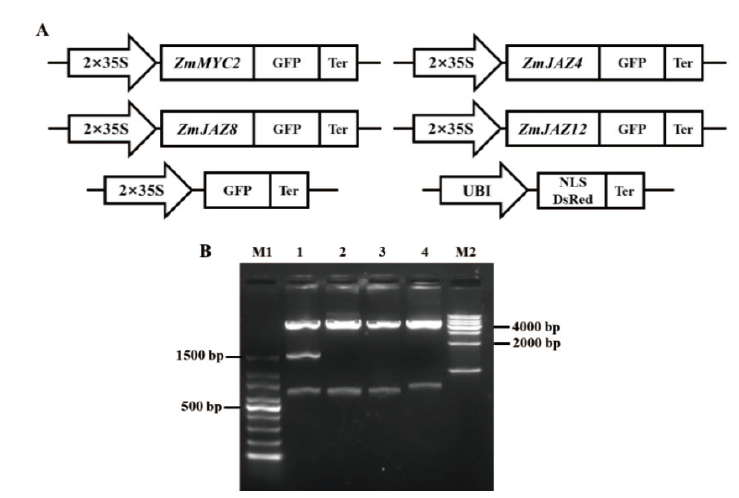
图2 亚细胞定位载体和酶切鉴定电泳图 A:亚细胞定位所用载体构建的示意图,包括对照GFP载体和细胞核定位marker载体。B:亚细胞定位载体的Xho I和Xba I酶切检测结果,其中M1为100 bp DNA Ladder;M2为1 kb DNA Ladder;泳道1为pZmMYC2-GFP;泳道2为pZmJAZ4-GFP;泳道3为pZmJAZ8-GFP;泳道4为pZmJAZ12-GFP
Fig.2 Vectors for subcellular localization and enzyme digestion identification A:Schematic diagram of vectors for subcellular localization,including control GFP vector and cell nucleus localization marker vector. B:Enzyme digestion identification of Xho I + Xba I in subcellular localization vectors. M1:100 bp DNA ladder. M2:1000 bp DNA ladder. Lane 1:pZmMYC2-GFP,lane 2:pZmJAZ4-GFP,lane 3:pZmJAZ8-GFP,and lane 4:pZmJAZ12-GFP
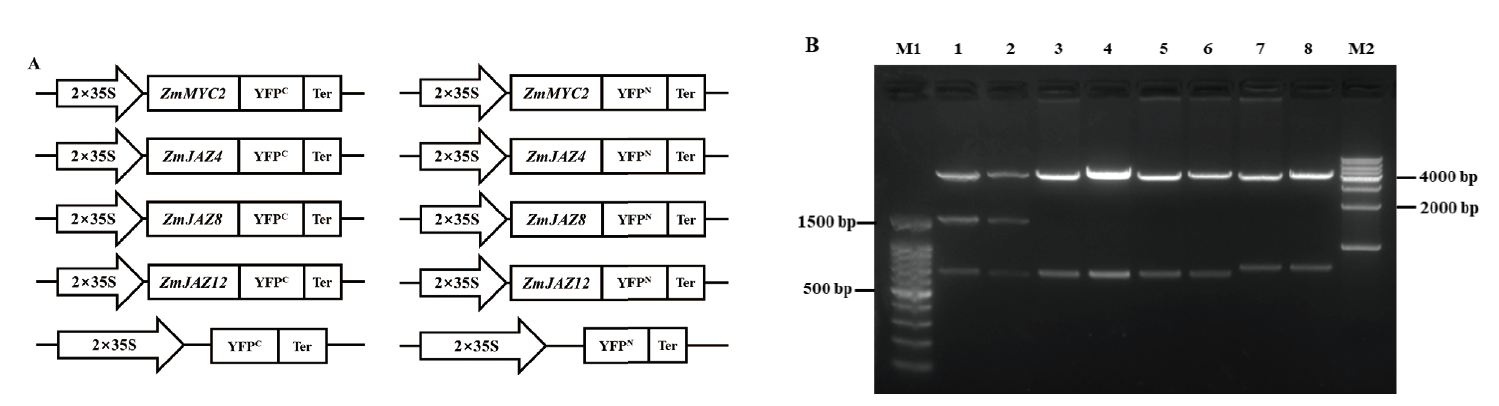
图3 BiFC载体构建示意图和酶切鉴定电泳图 A:BiFC所用载体构建的示意图。B:各BiFC载体的Xho I和Xba I酶切检测结果,其中M1为100 bp DNA Ladder;M2为全式金公司1 kb DNA Ladder;泳道1为pZmMYC2-YFPN;泳道2为pZmMYC2-YFPC;泳道3为pZmJAZ4-YFPN;泳道4为pZmJAZ4-YFPC;泳道5为pZmJAZ8-YFPN;泳道6为pZmJAZ8-YFPC;泳道7为pZmJAZ12-YFPN;泳道8为pZmJAZ12-YFPC
Fig.3 Vectors for BiFC assay and enzyme digestion identification A:Schematic diagram of vectors for BiFC assay. B:Enzyme digestion identification of BiFC vectors by Xho I + Xba I. M1:100 bp DNA ladder. M2:1 000 bp DNA ladder. Lane 1:pZmMYC2-YFPN,lane 2:pZmMYC2-YFPC,lane 3:pZmJAZ4-YFPN,lane 4:pZmJAZ4-YFPC,lane 5:pZmJAZ8-YFPN,lane 6:pZmJAZ8-YFPC,lane 7:pZmJAZ12-YFPN,and lane 8:pZmJAZ12-YFPC
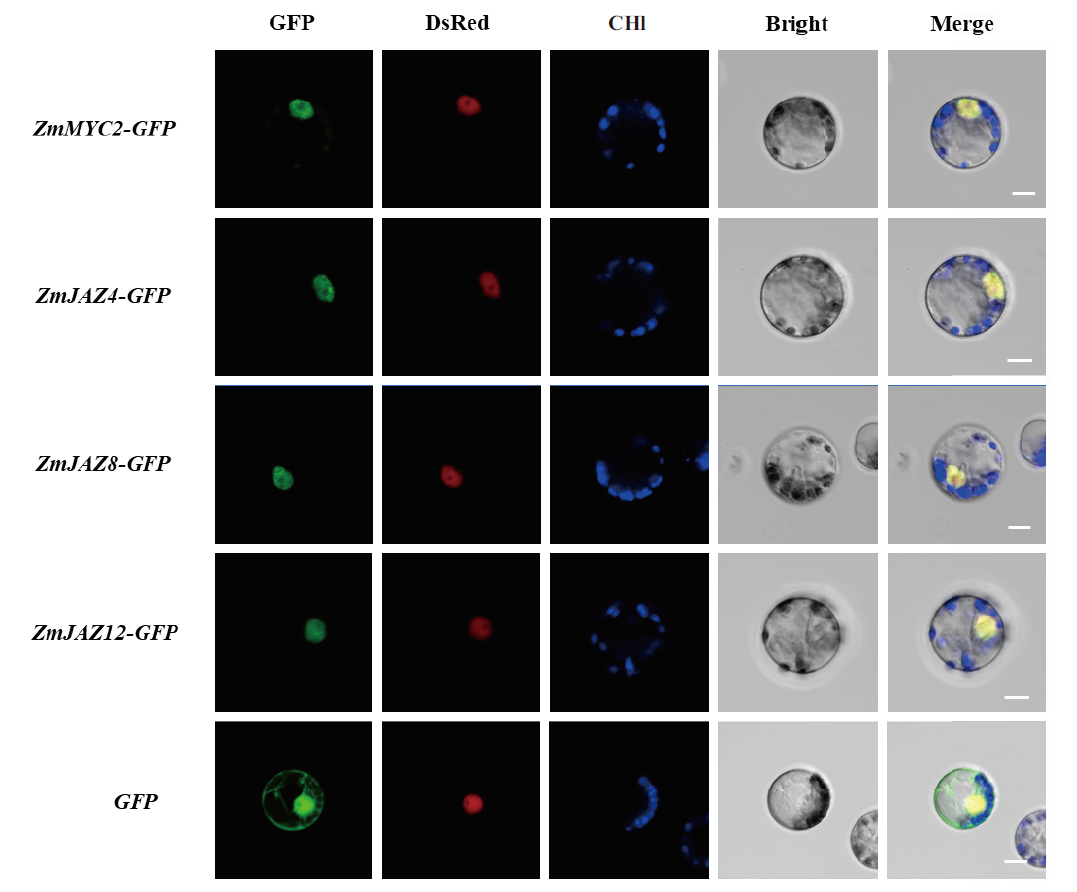
图4 ZmJAZ4、ZmJAZ8、ZmJAZ12和ZmMYC2的亚细胞定位 将ZmJAZ4、ZmJAZ8、ZmJAZ12和ZmMYC2构建到GFP的N端。GFP示目标蛋白荧光通道;DsRed示核定位标记载体荧光通道;Chl示叶绿体自发荧光通道;Bright示明场通道;Merge示GFP、DsRed、Chl和明场的叠加通道。Bar=10 μm
Fig. 4 Subcellular localization of ZmJAZ4,ZmJAZ8,ZmJAZ12 and ZmMYC2 The ORFs of ZmJAZ4、ZmJAZ8、ZmJAZ12 and ZmMYC2 were fused at the N terminal of GFP. GFP indicates the target protein fluorescence channel. DsRed indicates fluorescence channel for a nuclear localization maker. Chl indicates the chloroplast spontaneous fluorescence channel. Bright indicates the bright field. The Merge indicates channel combined the GFP,DsRed,Chl and the bright field. Scale bar = 10 μm

图5 ZmMYC2与ZmJAZ4、ZmJAZ8和ZmJAZ12的BiFC互作验证 将ZmMYC2和待验证的3个基因分别构建到YFP的N端及C端进行双向验证。YFP示BiFC互作激发荧光通道;DsRed示核定位信号载体荧光通道;Chl示叶绿体自发荧光通道;Bright示明场通道;Merge示YFP、DsRed、Chl和明场的叠加通道。Bar=10 μm
Fig. 5 BiFC assay to verify interactions between ZmJAZs with ZmMYC2 ZmMYC2 and 3 to-be-verified genes were constructed into the N-terminal and C-terminal of YFP for BiFC assay. YFP indicates fluorescence channel triggered by BiFC interaction. The DsRed indicates fluorescence channel for nuclear localization maker. Chl indicates the chloroplast spontaneous fluorescence channel. Bright indicates the bright field. Merge indicates channel combined the GFP,DsRed,Chl and the bright field. Scale bar = 10 μm
| [1] |
Wasternack C, Hause B. Jasmonates:biosynjournal, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany[J]. Ann Bot, 2013, 111(6):1021-1058.
doi: 10.1093/aob/mct067 URL |
| [2] |
Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant[J]. PNAS, 1992, 89(15):6837-6840.
pmid: 11607311 |
| [3] |
Berens ML, Berry HM, Mine A, et al. Evolution of hormone signaling networks in plant defense[J]. Annu Rev Phytopathol, 2017, 55:401-425.
doi: 10.1146/phyto.2017.55.issue-1 URL |
| [4] |
Huang H, Liu B, Liu L, et al. Jasmonate action in plant growth and development[J]. J Exp Bot, 2017, 68(6):1349-1359.
doi: 10.1093/jxb/erw495 pmid: 28158849 |
| [5] |
Dombrecht B, Xue GP, Sprague SJ, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis[J]. Plant Cell, 2007, 19(7):2225-2245.
pmid: 17616737 |
| [6] |
Chini A, Fonseca S, Fernández G, et al. The JAZ family of repressors is the missing link in jasmonate signalling[J]. Nature, 2007, 448(7154):666-671.
doi: 10.1038/nature06006 URL |
| [7] |
Yan J, Zhang C, Gu M, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor[J]. Plant Cell, 2009, 21(8):2220-2236.
doi: 10.1105/tpc.109.065730 URL |
| [8] |
Melotto M, Mecey C, Niu Y, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein[J]. Plant J, 2008, 55(6):979-988.
doi: 10.1111/tpj.2008.55.issue-6 URL |
| [9] |
Sheard LB, Tan X, Mao H, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor[J]. Nature, 2010, 468(7322):400-405.
doi: 10.1038/nature09430 URL |
| [10] |
Thines B, Katsir L, Melotto M, et al. JAZ repressor proteins are targets of the SCF(COI1)complex during jasmonate signalling[J]. Nature, 2007, 448(7154):661-665.
doi: 10.1038/nature05960 URL |
| [11] |
Zhang Z, Li X, Yu R, et al. Isolation, structural analysis, and expression characteristics of the maize TIFY gene family[J]. Mol Genet Genomics, 2015, 290(5):1849-1858.
doi: 10.1007/s00438-015-1042-6 URL |
| [12] |
Du M, Zhao J, Tzeng DTW, et al. MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato[J]. Plant Cell, 2017, 29(8):1883-1906.
doi: 10.1105/tpc.16.00953 URL |
| [13] |
Chen Q, Sun J, Zhai Q, et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis[J]. Plant Cell, 2011, 23(9):3335-3352.
doi: 10.1105/tpc.111.089870 URL |
| [14] |
Hong GJ, Xue XY, Mao YB, et al. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression[J]. Plant Cell, 2012, 24(6):2635-2648.
doi: 10.1105/tpc.112.098749 URL |
| [15] |
Fu J, Liu L, Liu Q, et al. ZmMYC2 exhibits diverse functions and enhances JA signaling in transgenic Arabidopsis[J]. Plant Cell Rep, 2020, 39(2):273-288.
doi: 10.1007/s00299-019-02490-2 URL |
| [16] |
Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses[J]. Plant Cell, 2011, 23(2):701-715.
doi: 10.1105/tpc.110.080788 URL |
| [17] |
Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis[J]. J Exp Bot, 2011, 62(6):2143-2154.
doi: 10.1093/jxb/erq408 URL |
| [18] |
Cheng Z, Sun L, Qi T, et al. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis[J]. Mol Plant, 2011, 4(2):279-288.
doi: 10.1093/mp/ssq073 URL |
| [19] |
Browse J, Wallis JG. Arabidopsis flowers unlocked the mechanism of jasmonate signaling[J]. Plants, 2019, 8(8):285.
doi: 10.3390/plants8080285 URL |
| [20] |
Wu H, Ye HY, Yao RF, et al. OsJAZ9 Acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice[J]. Plant Sci, 2015, 232:1-12.
doi: 10.1016/j.plantsci.2014.12.010 URL |
| [21] |
Acosta IF, Laparra H, Romero SP, et al. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize[J]. Science, 2009, 323(5911):262-265.
doi: 10.1126/science.1164645 pmid: 19131630 |
| [22] |
Xiao S, Dai LY, Liu FQ, et al. COS1:an Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence[J]. Plant Cell, 2004, 16(5):1132-1142.
pmid: 15075400 |
| [23] |
Shan XY, Wang JX, Chua L, et al. The role of Arabidopsis rubisco activase in jasmonate-induced leaf senescence[J]. Plant Physiol, 2011, 155(2):751-764.
doi: 10.1104/pp.110.166595 URL |
| [24] |
Feys B, Benedetti CE, Penfold CN, et al. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen[J]. Plant Cell, 1994, 6(5):751-759.
doi: 10.2307/3869877 URL |
| [25] |
Li L, Zhao Y, McCaig BC, et al. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development[J]. Plant Cell, 2004, 16(1):126-143.
doi: 10.1105/tpc.017954 URL |
| [26] |
晏胜伟, 孙程, 周晓今, 等. 玉米JAZ家族基因ZmJAZ4的克隆及功能分析[J]. 生物技术通报, 2015, 31(3):96-101.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.04.013 |
| Yan SW, Sun C, Zhou XJ, et al. Cloning and characterization analysis of zm JAZ4, a JAZ family gene in maize(Zea mays L.)[J]. Biotechnol Bull, 2015, 31(3):96-101. | |
| [27] |
Zheng J, Fu J, Gou M, et al. Genome-wide transcriptome analysis of two maize inbred lines under drought stress[J]. Plant Mol Biol, 2010, 72(4/5):407-421.
doi: 10.1007/s11103-009-9579-6 URL |
| [1] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [2] | 孙雨桐, 刘德帅, 齐迅, 冯美, 黄栩筝, 姚文孔. 茉莉酸调控植物生长发育和胁迫的研究进展[J]. 生物技术通报, 2023, 39(11): 99-109. |
| [3] | 张婵, 吴友根, 于靖, 杨东梅, 姚广龙, 杨华庚, 张军锋, 陈萍. 光与茉莉酸信号介导的萜类化合物合成分子机制[J]. 生物技术通报, 2022, 38(8): 32-40. |
| [4] | 韩志玲, 陈青, 梁晓, 伍春玲, 刘迎, 伍牧锋, 徐雪莲. 二斑叶螨取食抗、感螨木薯品种对茉莉酸信号途径基因表达的影响[J]. 生物技术通报, 2022, 38(6): 211-220. |
| [5] | 李治文, 刘培燕, 陈建松, 廖金铃, 林柏荣, 卓侃. 线虫效应子MgMO237及互作蛋白OsCRRSP55在水稻中的共响应基因鉴定[J]. 生物技术通报, 2021, 37(7): 88-97. |
| [6] | 钟李婷, 陈秀珍, 唐云, 李俊仁, 王小兵, 刘彦婷, 周璇璇, 詹若挺, 陈立凯. 广藿香FPPS重组蛋白表达及互作蛋白筛选分析[J]. 生物技术通报, 2019, 35(12): 10-15. |
| [7] | 梁东雨, 王禹佳, 苏敖纯, 徐洪伟, 周晓馥. 外源茉莉酸对牛皮杜鹃UV-B辐射缓解作用研究[J]. 生物技术通报, 2019, 35(10): 64-70. |
| [8] | 窦悦, 刘美彤, 卢安娜, 吴佳洁, 王群青, 胥倩. 中介体亚基MED25调控植物激素信号转导的研究进展[J]. 生物技术通报, 2018, 34(7): 40-47. |
| [9] | 熊丙全, 刘冬青, 廖相建, 郑雪莲. 茉莉酸甲酯对丹参毛状根有效成分含量的影响[J]. 生物技术通报, 2018, 34(7): 81-84. |
| [10] | 李杨洋, 焦浈. 外源茉莉酸甲酯对小麦幼苗低温耐受性的影响[J]. 生物技术通报, 2018, 34(3): 87-92. |
| [11] | 徐岩, 韩玉乾, 于放, 刘志文, 王燕燕. 过表达长春花JAR1基因促进文朵灵和长春质碱的生物合成[J]. 生物技术通报, 2017, 33(6): 62-68. |
| [12] | 邹广平, 李雅丽, 冯梦薇, 杨修, 许智伟. 甘草悬浮细胞肉桂酸-4-羟基化酶(C4H)基因的克隆及表达分析[J]. 生物技术通报, 2017, 33(11): 96-100. |
| [13] | 柳琳, 彭波, 蒋春苗, 程在全. 疣粒野生稻胚性细胞与叶片抗白叶枯病信号物质和相关抗病基因的表达差异[J]. 生物技术通报, 2016, 32(10): 199-204. |
| [14] | 潘益芳, 龚一富, 俞凯, 朱帅旗, 王何瑜. 绿色杜氏藻DHDDS基因生物信息学分析及表达研究[J]. 生物技术通报, 2015, 31(7): 101-108. |
| [15] | 郭航, 王志敏, 汤青林, 田时炳, 杨洋 , 宋明. 茉莉酸调控花药开裂的研究进展[J]. 生物技术通报, 2015, 31(2): 10-17. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||