








生物技术通报 ›› 2022, Vol. 38 ›› Issue (2): 10-20.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0542
李兵娟1,2( ), 郑璐1, 沈仁芳1,2, 兰平1,2(
), 郑璐1, 沈仁芳1,2, 兰平1,2( )
)
收稿日期:2021-04-23
出版日期:2022-02-26
发布日期:2022-03-09
作者简介:李兵娟,女,博士研究生,研究方向:植物营养蛋白质组学;E-mail: 基金资助:
LI Bing-juan1,2( ), ZHENG Lu1, SHEN Ren-fang1,2, LAN Ping1,2(
), ZHENG Lu1, SHEN Ren-fang1,2, LAN Ping1,2( )
)
Received:2021-04-23
Published:2022-02-26
Online:2022-03-09
摘要:
核糖体蛋白不仅参与蛋白质合成,而且参与植物生长发育的调控。利用拟南芥核糖体磷酸蛋白P1(ribosomal phosphoprotein P1,RPP1)家族基因RPP1A缺失突变体rpp1a研究RPP1A缺失对幼苗蛋白质表达水平的影响,揭示其参与调控幼苗生长的作用机制。表型分析发现,与野生型WT相比,RPP1A缺失导致拟南芥幼苗生物量、叶片总表面积和叶柄长度显著增加。基于label-free的蛋白质组学分析,与野生型相比,rpp1a-2突变体有280个蛋白质表达量具有显著差异,其中98个显著上调,182个显著下调。差异蛋白主要富集在细胞过程、代谢过程、刺激反应、细胞成分组织或生物发生和氧化还原过程等生物学过程,并在核糖体、光合作用、mRNA监视途径、RNA降解和苯丙素的生物合成代谢通路中显著富集。拟南芥rpp1a-2突变体通过抑制刺激反应和增强光合作用等过程促进植物生长。
李兵娟, 郑璐, 沈仁芳, 兰平. 拟南芥RPP1A参与幼苗生长的蛋白质组学分析[J]. 生物技术通报, 2022, 38(2): 10-20.
LI Bing-juan, ZHENG Lu, SHEN Ren-fang, LAN Ping. Proteomic Analysis of RPP1A Involved in the Seedling Growth of Arabidopsis thaliana[J]. Biotechnology Bulletin, 2022, 38(2): 10-20.
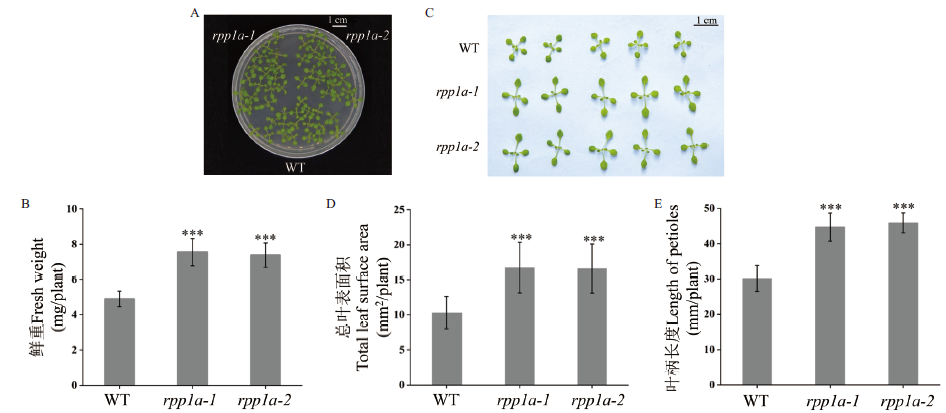
图1 拟南芥WT和rpp1a-1和rpp1a-2突变体幼苗表型 拟南芥WT和rpp1a-1和rpp1a-2突变体在ES培养基上生长14 d的幼苗表型(A)和幼苗鲜重(B)、叶片表型(C)、叶片总表面积(D)和叶柄长度(E)。数值以±s差表示,采用one-way ANOVA检验进行显著性分析,***表示极显著差异(P<0.001)
Fig. 1 Seedling phenotypes of Arabidopsis thaliana WT and rpp1a-1 and rpp1a-2 mutants Phenotype of seedlings(A),fresh weight(B),leaf phenotype(C),total leaf surface area(D),and length of petioles(E)of Arabidopsis thaliana WT and rpp1a-1 and rpp1a-2 mutants growing on ES medium for 14 d. Values are expressed as ±s deviation,and a one-way ANOVA test is used for significance analysis. *** refers to extremely significant difference(P<0.001)
| 拟南芥基因号 AGI locus ID | 蛋白质名称 Protein name | 描述 Description | 丰度比Abundance ratio | 丰度比的调整P值 Abundance ratio adj. P value | 丰度比变异性 Abundance ratio variability/% |
|---|---|---|---|---|---|
| 核糖体Ribosome | |||||
| AT3G49080 | RPS9M | 核糖体蛋白S9 M Ribosomal protein S9 M | 100.00 | 2.02E-16 | 0.00 |
| AT2G21580 | RPS25B | 核糖体蛋白S25家族蛋白 Ribosomal protein S25 family protein | 11.39 | 2.02E-16 | 642.26 |
| AT2G19740 | RPL31A | 核糖体蛋白L31e家族蛋白 Ribosomal protein L31e family protein | 0.63 | 3.50E-02 | 7.21 |
| AT5G56670 | RPS30C | 核糖体蛋白S30家族蛋白 Ribosomal protein S30 family protein | 0.59 | 4.08E-02 | 3.33 |
| AT3G46040 | RPS15aD | 核糖体蛋白S15a D Ribosomal protein S15a D | 0.58 | 2.65E-02 | 1.03 |
| AT5G16130 | RPS7C | 核糖体蛋白S7e家族蛋白 Ribosomal protein S7e family protein | 0.57 | 8.83E-04 | 2.09 |
| AT1G04270 | RPS15A | 核糖体蛋白S15 Ribosomal protein S15 | 0.53 | 6.16E-03 | 14.83 |
| AT5G18380 | RPS16C | 核糖体蛋白S5结构域2类似超家族蛋白Ribosomal protein S5 domain 2-like superfamily protein | 0.51 | 1.27E-04 | 8.10 |
| AT5G23900 | RPL13D | 核糖体蛋白L13e家族蛋白 Ribosomal protein L13e family protein | 0.49 | 1.49E-04 | 4.12 |
| AT2G27720 | RPP2A | 60S酸性核糖体蛋白家族 60S acidic ribosomal protein family | 0.43 | 1.14E-05 | 1.49 |
| AT3G28900 | RPL34C | 核糖体蛋白L34e超家族蛋白质 Ribosomal protein L34e superfamily protein | 0.33 | 1.77E-04 | 2.55 |
| AT1G01100 | RPP1A | 60S酸性核糖体蛋白家族 60S acidic ribosomal protein family | 0.01 | 2.02E-16 | 0.00 |
| 光合作用Photosynthesis | |||||
| ATCG00020 | PSBA | 光系统II反应中心蛋白D1 Phot-osystem II reaction center protein D1 | 1.75 | 6.17E-04 | 18.27 |
| ATCG00270 | PSBD | P光系统II反应中心蛋白D2 Pho-tosystem II reaction center protein D2 | 1.69 | 1.53E-03 | 16.06 |
| ATCG00580 | PSBE | 光系统II反应中心蛋白V Phot-osystem II reaction center protein V | 0.62 | 3.90E-02 | 7.74 |
| ATCG00470 | ATPE | ATP合酶ε亚基 ATP synthase epsilon subunit | 0.58 | 1.20E-02 | 4.17 |
| AT1G10960 | FD1 | 铁氧还原蛋白1 Ferredoxin 1 | 0.01 | 2.02E-16 | 0.00 |
| mRNA监视通路mRNA surveillance pathway | |||||
| AT1G13320 | PP2AA3 | 蛋白磷酸酶2A亚基A3 Protein phosphatase 2A subunit A3 | 100.00 | 2.02E-16 | 0.00 |
| AT1G02140 | MAGO | Mago nashi家族蛋白 Mago nashi family protein | 2.23 | 1.48E-05 | 20.68 |
| AT1G17720 | ATB BETA | 蛋白磷酸酶2A,调控亚基PR55 Protein phosphatase 2A,regulatory subunit PR55 | 0.13 | 1.44E-12 | 15.32 |
| AT1G16610 | SR45 | 精氨酸/丝氨酸丰富45 Arginine/serine-rich 45 | 0.06 | 2.02E-16 | 1.89 |
| AT5G51660 | CPSF160 | 切割与多聚腺苷酸化特异性因子160 Cleavage and polyadenylation specificity factor 160 | 0.01 | 2.02E-16 | 0.00 |
| AT5G47010 | LBA1 | RNA解旋酶 RNA helicase | 0.01 | 2.02E-16 | 0.00 |
| RNA降解RNA degradation | |||||
| AT5G61580 | PFK4 | 磷酸果糖激酶4 Phosphofructokinase 4 | 100.00 | 2.02E-16 | 0.00 |
| AT5G02270 | ABCI20 | 非本征ABC蛋白9 Non-intrinsic ABC protein 9 | 0.46 | 2.02E-16 | 4.33 |
| AT5G35430 | NOT10 | 四肽重复序列类超家族蛋白 Tetratricopeptide repeat(TPR)-like superfamily protein | 0.04 | 2.02E-16 | 0.36 |
| AT4G32840 | PFK6 | 磷酸果糖激酶6 Phosphofructokinase 6 | 0.01 | 2.02E-16 | 0.00 |
| AT3G20800 | NOT9A | 细胞分化,Rcd1类蛋白 Cell differentiation,Rcd1-like protein | 0.01 | 8.72E-03 | 0.00 |
| 苯丙素的生物合成Phenylpropanoid biosynthesis | |||||
| AT2G18980 | PRX16 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 100.00 | 2.02E-16 | 0.00 |
| AT5G66390 | PRX72 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 2.07 | 2.02E-02 | 41.96 |
| AT1G66270 | BGLU21 | 糖基水解酶超家族蛋白,β-葡萄糖苷酶 Glycosyl hydrolase superfamily protein,β-glucosidase | 0.63 | 4.18E-02 | 1.71 |
| AT1G66280 | BGLU22 | 糖基水解酶超家族蛋白,β-葡萄糖苷酶 Glycosyl hydrolase superfamily protein,β-glucosidase | 0.44 | 3.47E-06 | 7.12 |
| AT5G64120 | PRX71 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 0.30 | 1.57E-11 | 1.14 |
| AT2G38390 | PRX23 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 0.14 | 3.52E-14 | 0.75 |
| AT4G33420 | PRX47 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 0.11 | 1.57E-11 | 9.98 |
表1 显著富集的KEGG通路中的差异表达蛋白
Table 1 Differentially expressed proteins in the significantly enriched KEGG pathways
| 拟南芥基因号 AGI locus ID | 蛋白质名称 Protein name | 描述 Description | 丰度比Abundance ratio | 丰度比的调整P值 Abundance ratio adj. P value | 丰度比变异性 Abundance ratio variability/% |
|---|---|---|---|---|---|
| 核糖体Ribosome | |||||
| AT3G49080 | RPS9M | 核糖体蛋白S9 M Ribosomal protein S9 M | 100.00 | 2.02E-16 | 0.00 |
| AT2G21580 | RPS25B | 核糖体蛋白S25家族蛋白 Ribosomal protein S25 family protein | 11.39 | 2.02E-16 | 642.26 |
| AT2G19740 | RPL31A | 核糖体蛋白L31e家族蛋白 Ribosomal protein L31e family protein | 0.63 | 3.50E-02 | 7.21 |
| AT5G56670 | RPS30C | 核糖体蛋白S30家族蛋白 Ribosomal protein S30 family protein | 0.59 | 4.08E-02 | 3.33 |
| AT3G46040 | RPS15aD | 核糖体蛋白S15a D Ribosomal protein S15a D | 0.58 | 2.65E-02 | 1.03 |
| AT5G16130 | RPS7C | 核糖体蛋白S7e家族蛋白 Ribosomal protein S7e family protein | 0.57 | 8.83E-04 | 2.09 |
| AT1G04270 | RPS15A | 核糖体蛋白S15 Ribosomal protein S15 | 0.53 | 6.16E-03 | 14.83 |
| AT5G18380 | RPS16C | 核糖体蛋白S5结构域2类似超家族蛋白Ribosomal protein S5 domain 2-like superfamily protein | 0.51 | 1.27E-04 | 8.10 |
| AT5G23900 | RPL13D | 核糖体蛋白L13e家族蛋白 Ribosomal protein L13e family protein | 0.49 | 1.49E-04 | 4.12 |
| AT2G27720 | RPP2A | 60S酸性核糖体蛋白家族 60S acidic ribosomal protein family | 0.43 | 1.14E-05 | 1.49 |
| AT3G28900 | RPL34C | 核糖体蛋白L34e超家族蛋白质 Ribosomal protein L34e superfamily protein | 0.33 | 1.77E-04 | 2.55 |
| AT1G01100 | RPP1A | 60S酸性核糖体蛋白家族 60S acidic ribosomal protein family | 0.01 | 2.02E-16 | 0.00 |
| 光合作用Photosynthesis | |||||
| ATCG00020 | PSBA | 光系统II反应中心蛋白D1 Phot-osystem II reaction center protein D1 | 1.75 | 6.17E-04 | 18.27 |
| ATCG00270 | PSBD | P光系统II反应中心蛋白D2 Pho-tosystem II reaction center protein D2 | 1.69 | 1.53E-03 | 16.06 |
| ATCG00580 | PSBE | 光系统II反应中心蛋白V Phot-osystem II reaction center protein V | 0.62 | 3.90E-02 | 7.74 |
| ATCG00470 | ATPE | ATP合酶ε亚基 ATP synthase epsilon subunit | 0.58 | 1.20E-02 | 4.17 |
| AT1G10960 | FD1 | 铁氧还原蛋白1 Ferredoxin 1 | 0.01 | 2.02E-16 | 0.00 |
| mRNA监视通路mRNA surveillance pathway | |||||
| AT1G13320 | PP2AA3 | 蛋白磷酸酶2A亚基A3 Protein phosphatase 2A subunit A3 | 100.00 | 2.02E-16 | 0.00 |
| AT1G02140 | MAGO | Mago nashi家族蛋白 Mago nashi family protein | 2.23 | 1.48E-05 | 20.68 |
| AT1G17720 | ATB BETA | 蛋白磷酸酶2A,调控亚基PR55 Protein phosphatase 2A,regulatory subunit PR55 | 0.13 | 1.44E-12 | 15.32 |
| AT1G16610 | SR45 | 精氨酸/丝氨酸丰富45 Arginine/serine-rich 45 | 0.06 | 2.02E-16 | 1.89 |
| AT5G51660 | CPSF160 | 切割与多聚腺苷酸化特异性因子160 Cleavage and polyadenylation specificity factor 160 | 0.01 | 2.02E-16 | 0.00 |
| AT5G47010 | LBA1 | RNA解旋酶 RNA helicase | 0.01 | 2.02E-16 | 0.00 |
| RNA降解RNA degradation | |||||
| AT5G61580 | PFK4 | 磷酸果糖激酶4 Phosphofructokinase 4 | 100.00 | 2.02E-16 | 0.00 |
| AT5G02270 | ABCI20 | 非本征ABC蛋白9 Non-intrinsic ABC protein 9 | 0.46 | 2.02E-16 | 4.33 |
| AT5G35430 | NOT10 | 四肽重复序列类超家族蛋白 Tetratricopeptide repeat(TPR)-like superfamily protein | 0.04 | 2.02E-16 | 0.36 |
| AT4G32840 | PFK6 | 磷酸果糖激酶6 Phosphofructokinase 6 | 0.01 | 2.02E-16 | 0.00 |
| AT3G20800 | NOT9A | 细胞分化,Rcd1类蛋白 Cell differentiation,Rcd1-like protein | 0.01 | 8.72E-03 | 0.00 |
| 苯丙素的生物合成Phenylpropanoid biosynthesis | |||||
| AT2G18980 | PRX16 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 100.00 | 2.02E-16 | 0.00 |
| AT5G66390 | PRX72 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 2.07 | 2.02E-02 | 41.96 |
| AT1G66270 | BGLU21 | 糖基水解酶超家族蛋白,β-葡萄糖苷酶 Glycosyl hydrolase superfamily protein,β-glucosidase | 0.63 | 4.18E-02 | 1.71 |
| AT1G66280 | BGLU22 | 糖基水解酶超家族蛋白,β-葡萄糖苷酶 Glycosyl hydrolase superfamily protein,β-glucosidase | 0.44 | 3.47E-06 | 7.12 |
| AT5G64120 | PRX71 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 0.30 | 1.57E-11 | 1.14 |
| AT2G38390 | PRX23 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 0.14 | 3.52E-14 | 0.75 |
| AT4G33420 | PRX47 | 过氧化物酶超家族蛋白 Peroxidase superfamily protein | 0.11 | 1.57E-11 | 9.98 |
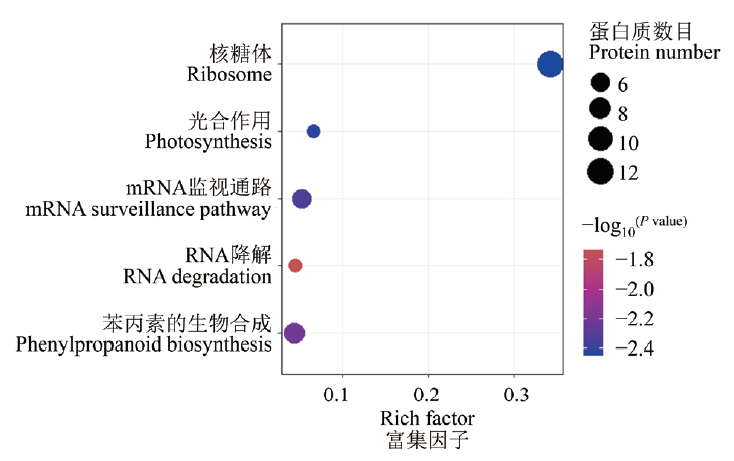
图5 WT和rpp1a-2突变体幼苗差异蛋白的KEGG富集分析
Fig. 5 KEGG enrichment analysis of differentially expr-essed proteins identified in the WT and rpp1a-2 mutant seedlings
图6 差异蛋白的蛋白质互作网络 圆圈大小与互作关系的数目成正比
Fig. 6 Protein-protein interaction network of differentially expressed proteins Size of the circle is proportional to the number of interaction relationships
| [1] | 靳聪聪, 侯名语, 潘延云. 拟南芥核糖体蛋白生物学功能研究进展[J]. 植物生理学报, 2018, 54(2):203-212. |
| Jin CC, Hou MY, et al. Research progress of ribosomal protein function in Arabidopsis thaliana[J]. Plant Physiol J, 2018, 54(2):203-212. | |
| [2] |
Weis BL, Kovacevic J, et al. Plant-specific features of ribosome biogenesis[J]. Trends Plant Sci, 2015, 20(11):729-740.
doi: 10.1016/j.tplants.2015.07.003 URL |
| [3] |
Barakat A, Szick-Miranda K, Chang IF, et al. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome[J]. Plant Physiol, 2001, 127(2):398-415.
pmid: 11598216 |
| [4] |
Carroll AJ, Heazlewood JL, Ito J, et al. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification[J]. Mol Cell Proteomics, 2008, 7(2):347-369.
pmid: 17934214 |
| [5] |
Hummel M, Dobrenel T, Cordewener JJ, et al. Proteomic LC-MS analysis of Arabidopsis cytosolic ribosomes:Identification of ribosomal protein paralogs and re-annotation of the ribosomal protein genes[J]. J Proteomics, 2015, 128:436-449.
doi: 10.1016/j.jprot.2015.07.004 URL |
| [6] |
Savada RP, Bonham-Smith PC. Differential transcript accumulation and subcellular localization of Arabidopsis ribosomal proteins[J]. Plant Sci, 2014, 223:134-145.
doi: 10.1016/j.plantsci.2014.03.011 URL |
| [7] |
Wang J, Lan P, Gao H, et al. Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition[J]. BMC Genomics, 2013, 14:783.
doi: 10.1186/1471-2164-14-783 URL |
| [8] |
Horiguchi G, Mollá-Morales A, Pérez-Pérez JM, et al. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development[J]. Plant J, 2011, 65(5):724-736.
doi: 10.1111/tpj.2011.65.issue-5 URL |
| [9] | Norris K, Hopes T, Aspden JL. Ribosome heterogeneity and specialization in development[J]. Wiley Interdiscip Rev RNA, 2021: e1644. |
| [10] |
Fernández-Pérez F, Pomar F, Pedreño MA, et al. Suppression of Arabidopsis peroxidase 72 alters cell wall and phenylpropanoid metabolism[J]. Plant Sci, 2015, 239:192-199.
doi: 10.1016/j.plantsci.2015.08.001 pmid: 26398803 |
| [11] |
Szakonyi D, Byrne ME. Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana[J]. Plant J, 2011, 65(2):269-281.
doi: 10.1111/tpj.2011.65.issue-2 URL |
| [12] |
Yan H, Chen D, Wang Y, et al. Ribosomal protein L18aB is required for both male gametophyte function and embryo development in Arabidopsis[J]. Sci Rep, 2016, 6:31195.
doi: 10.1038/srep31195 URL |
| [13] |
Ramos RS, Casati P, et al. Ribosomal protein RPL10A contributes to early plant development and abscisic acid-dependent responses in Arabidopsis[J]. Front Plant Sci, 2020, 11:582353.
doi: 10.3389/fpls.2020.582353 URL |
| [14] |
Falcone Ferreyra ML, Pezza A, Biarc J, et al. Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress[J]. Plant Physiol, 2010, 153(4):1878-1894.
doi: 10.1104/pp.110.157057 pmid: 20516338 |
| [15] |
Jorrín-Novo JV, Pascual J, Sánchez-Lucas R, et al. Fourteen years of plant proteomics reflected in Proteomics:moving from model species and 2DE-based approaches to orphan species and gel-free platforms[J]. Proteomics, 2015, 15(5/6):1089-1112.
doi: 10.1002/pmic.201400349 URL |
| [16] | 王立赛, 闫明科, 王晗, 等. 一个响应土壤缺铁拟南芥突变体的分离及鉴定[J]. 土壤, 2018, 50(3):476-484. |
| Wang LS, Yan MK, Wang H, et al. Screening and characterization of an Arabidopsis mutant in response to iron deficiency[J]. Soils, 2018, 50(3):476-484. | |
| [17] |
Lan P, Li W, Schmidt W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation[J]. Mol Cell Proteomics, 2012, 11(11):1156-1166.
doi: 10.1074/mcp.M112.020461 URL |
| [18] |
Wiśniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis[J]. Nat Methods, 2009, 6(5):359-362.
doi: 10.1038/nmeth.1322 pmid: 19377485 |
| [19] |
Yan M, Zheng L, Li B, et al. Comparative proteomics reveals new insights into the endosperm responses to drought, salinity and submergence in germinating wheat seeds[J]. Plant Mol Biol, 2021, 105(3):287-302.
doi: 10.1007/s11103-020-01087-8 URL |
| [20] | Cheng CY, Krishnakumar V, Chan A, et al. Araport11:a complete reannotation of the Arabidopsis thaliana reference genome[J]. Plant, 2017, 89(4):789-804. |
| [21] |
Tian T, Liu Y, Yan H, et al. agriGO v2. 0:a GO analysis toolkit for the agricultural community, 2017 update[J]. Nucleic Acids Res, 2017, 45(w1):W122-W129.
doi: 10.1093/nar/gkx382 URL |
| [22] |
Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets[J]. Nat Commun, 2019, 10(1):1523.
doi: 10.1038/s41467-019-09234-6 URL |
| [23] |
Carroll AJ. The Arabidopsis cytosolic ribosomal proteome:from form to function[J]. Front Plant Sci, 2013, 4:32.
doi: 10.3389/fpls.2013.00032 pmid: 23459595 |
| [24] |
Zsögön A, Szakonyi D, Shi XL, et al. Ribosomal protein RPL27a promotes female gametophyte development in a dose-dependent manner[J]. Plant Physiol, 2014, 165(3):1133-1143.
doi: 10.1104/pp.114.241778 URL |
| [25] |
Xiong W, Chen XZ, Zhu CX, et al. Arabidopsis paralogous genes RPL23aA and RPL23aB encode functionally equivalent proteins[J]. BMC Plant Biol, 2020, 20(1):463.
doi: 10.1186/s12870-020-02672-1 pmid: 33032526 |
| [26] |
Yang M, Wang X, et al. Genomic architecture of biomass heterosis in Arabidopsis[J]. PNAS, 2017, 114(30):8101-8106.
doi: 10.1073/pnas.1705423114 pmid: 28696287 |
| [27] |
Prasinos C, Krampis K, Samakovli D, et al. Tight regulation of expression of two Arabidopsis cytosolic Hsp90 genes during embryo development[J]. J Exp Bot, 2005, 56(412):633-644.
doi: 10.1093/jxb/eri035 URL |
| [28] |
Yang Y, Karlson D. AtCSP1 regulates germination timing promoted by low temperature[J]. FEBS Lett, 2013, 587(14):2186-2192.
doi: 10.1016/j.febslet.2013.05.039 URL |
| [29] |
Balkunde R, Foroughi L, Ewan E, et al. Mechanism of microtubule plus-end tracking by the plant-specific SPR1 protein and its development as a versatile plus-end marker[J]. J Biol Chem, 2019, 294(44):16374-16384.
doi: 10.1074/jbc.RA119.008866 URL |
| [30] |
Guo D, Gao X, Li H, et al. EGY1 plays a role in regulation of endodermal plastid size and number that are involved in ethylene-dependent gravitropism of light-grown Arabidopsis hypocotyls[J]. Plant Mol Biol, 2008, 66(4):345-360.
doi: 10.1007/s11103-007-9273-5 pmid: 18097640 |
| [31] |
Lu C, Yu F, Tian L, et al. RPS9M, a mitochondrial ribosomal protein, is essential for central cell maturation and endosperm development in Arabidopsis[J]. Front Plant Sci, 2017, 8:2171.
doi: 10.3389/fpls.2017.02171 URL |
| [32] |
Bach-Pages M, Homma F, et al. Discovering the RNA-binding proteome of plant leaves with an improved RNA interactome capture method[J]. Biomolecules, 2020, 10(4):661.
doi: 10.3390/biom10040661 URL |
| [33] |
Johnson MP. Photosynjournal[J]. Essays Biochem, 2016, 60(3):255-273.
doi: 10.1042/EBC20160016 URL |
| [34] |
Mulo P, Sirpiö S, Suorsa M, et al. Auxiliary proteins involved in the assembly and sustenance of photosystem II[J]. Photosynth Res, 2008, 98(1/2/3):489-501.
doi: 10.1007/s11120-008-9320-3 URL |
| [35] | Jin H, Fu M, Duan Z, et al. LOW PHOTOSYNTHETIC EFFICIENCY 1 is required for light-regulated photosystem II biogenesis in Arabidopsis[J]. PNAS, 2018, 115(26):E6075-E6084. |
| [36] |
Tokunaga N, Kaneta T, et al. Analysis of expression profiles of three peroxidase genes associated with lignification in Arabidopsis thaliana[J]. Physiol Plant, 2009, 136(2):237-249.
doi: 10.1111/j.1399-3054.2009.01233.x pmid: 19453502 |
| [37] |
Raggi S, Ferrarini A, Delledonne M, et al. The Arabidopsis class III peroxidase AtPRX71 negatively regulates growth under physiological conditions and in response to cell wall damage[J]. Plant Physiol, 2015, 169(4):2513-2525.
doi: 10.1104/pp.15.01464 pmid: 26468518 |
| [38] |
Shigeto J, Itoh Y, et al. Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem[J]. J Integr Plant Biol, 2015, 57(4):349-356.
doi: 10.1111/jipb.12334 pmid: 25644691 |
| [39] |
Vanholme R, De Meester B, Ralph J, et al. Lignin biosynjournal and its integration into metabolism[J]. Curr Opin Biotechnol, 2019, 56:230-239.
doi: 10.1016/j.copbio.2019.02.018 URL |
| [40] |
Vogt T. Phenylpropanoid biosynjournal[J]. Mol Plant, 2010, 3(1):2-20.
doi: 10.1093/mp/ssp106 URL |
| [41] |
Park NI, Yeung EC, Muench DG. Mago Nashi is involved in meristem organization, pollen formation, and seed development in Arabidopsis[J]. Plant Sci, 2009, 176(4):461-469.
doi: 10.1016/j.plantsci.2008.12.016 URL |
| [1] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [2] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [3] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [4] | 罗义, 张丽娟, 黄伟, 王宁, 吾尔丽卡·买提哈斯木, 施宠, 王玮. 一株耐铀菌株的鉴定及其促生特性研究[J]. 生物技术通报, 2023, 39(5): 286-296. |
| [5] | 王琪, 胡哲, 富薇, 李光哲, 郝林. 伯克霍尔德氏菌GD17对黄瓜幼苗耐干旱的调节[J]. 生物技术通报, 2023, 39(3): 163-175. |
| [6] | 崔吉洁, 蔡文波, 庄庆辉, 高爱平, 黄建峰, 陈亚辉, 宋志忠. 杧果Fe-S簇装配基因MiISU1的生物学功能[J]. 生物技术通报, 2023, 39(2): 139-146. |
| [7] | 鄢梦雨, 韦晓薇, 曹婧, 兰海燕. 异子蓬SabHLH169基因的克隆及抗旱功能分析[J]. 生物技术通报, 2023, 39(11): 328-339. |
| [8] | 阮航, 多浩源, 范文艳, 吕清晗, 姜述君, 朱生伟. AtERF49在拟南芥应答盐碱胁迫中的作用[J]. 生物技术通报, 2023, 39(1): 150-156. |
| [9] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [10] | 高聪, 萧楚健, 鲁帅, 王苏蓉, 袁卉华, 曹云英. 氧化石墨烯对拟南芥生长的促进作用[J]. 生物技术通报, 2022, 38(6): 120-128. |
| [11] | 徐红云, 张明意. GRAS转录因子AtSCL4负调控拟南芥应答渗透胁迫[J]. 生物技术通报, 2022, 38(6): 129-135. |
| [12] | 古盼, 齐学影, 李莉, 张曦, 单晓昳. AtRGS1胞吞动态调控G蛋白参与拟南芥生长发育和抗性反应[J]. 生物技术通报, 2022, 38(6): 34-42. |
| [13] | 赵明明, 唐殷, 郭磊周, 韩佳慧, 葛佳茗, 孟勇, 平淑珍, 周正富, 王劲. Lon1蛋白酶参与耐辐射异常球菌高温胁迫及细胞分裂的功能研究[J]. 生物技术通报, 2022, 38(5): 149-158. |
| [14] | 祖国蔷, 胡哲, 王琪, 李光哲, 郝林. Burkholderia sp. GD17对水稻幼苗镉耐受的调节[J]. 生物技术通报, 2022, 38(4): 153-162. |
| [15] | 周娟, 阎晋东, 李新梅, 刘雪晴, 赵强, 赵小英. 拟南芥F-box蛋白FKF1与转录因子FUL互作调控开花研究[J]. 生物技术通报, 2022, 38(3): 1-8. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||