








生物技术通报 ›› 2022, Vol. 38 ›› Issue (3): 121-129.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0471
收稿日期:2021-04-11
出版日期:2022-03-26
发布日期:2022-04-06
作者简介:付雅丽,女,硕士研究生,研究方向:生物化学与分子生物学;E-mail: 基金资助:
FU Ya-li( ), PENG Wan-li, LIN Shuang-jun, DENG Zi-xin, LIANG Ru-bing(
), PENG Wan-li, LIN Shuang-jun, DENG Zi-xin, LIANG Ru-bing( )
)
Received:2021-04-11
Published:2022-03-26
Online:2022-04-06
摘要:
香茅醇假单胞菌SJTE-3能够以17β-雌二醇为唯一碳源并将其高效降解,但其催化雌二醇转化的关键酶仍不明确。本文鉴定了该菌株中降解雌二醇的短链脱氢酶SDR-X1(WP_043267487.1),并对其功能进行了研究。首先利用荧光定量PCR,检测了不同碳源条件下基因sdr-x1的转录水平;克隆基因sdr-x1,在大肠杆菌BL21(DE3)菌株中诱导表达,利用亲和层析纯化获得了重组蛋白SDR-X1;体外检测了重组蛋白SDR-X1的催化活性与酶学性质,并利用高效液相色谱鉴定了其转化17β-雌二醇的产物。蛋白SDR-X1可被17β-雌二醇诱导表达;蛋白序列比对显示蛋白SDR-X1含有短链脱氢酶的保守基序与氨基酸。该酶以NAD+为辅因子,将17β-雌二醇氧化为雌酮;其Km值为(0.039 86±0.004 061)mmol/L,Vmax值为(3.168±0.135)mmol/L/min/mg,可在15 min内转化61.75%以上的雌二醇。该酶对温度具有一定耐受性,最佳反应温度为50℃,偏碱性pH可促进其酶促反应。不同二价金属离子对该酶活性具有不同的影响,Mg2+、Mn2+和Ca2+可增强其酶活性。香茅醇假单胞菌SJTE-3中的SDR-X1可高效催化17β-雌二醇转化,参与该菌株的雌激素降解过程,其功能研究将推进细菌的雌激素代谢机制解析。
付雅丽, 彭万里, 林双君, 邓子新, 梁如冰. 香茅醇假单胞菌SJTE-3的短链脱氢酶SDR-X1的克隆及酶性质测定[J]. 生物技术通报, 2022, 38(3): 121-129.
FU Ya-li, PENG Wan-li, LIN Shuang-jun, DENG Zi-xin, LIANG Ru-bing. Gene Cloning and Enzymatic Properties of the Short Chain Dehydrogenase SDR-X1 from Pseudomonas citronellolis SJTE-3[J]. Biotechnology Bulletin, 2022, 38(3): 121-129.

图1 蛋白SDR-X1的多序列比对、高级结构模型与进化分析 A:不同微生物来源的短链脱氢酶序列比对。包括来自香茅醇假单胞菌SJTE-3的蛋白SDR-X1(WP_043267487.1),欧洲亚硝化单胞菌(Nitrosomonas europaea)ATCC19718的蛋白CAD85559.1,赤红球菌(Rhodococcus ruber)P14的蛋白WP_010595991.1和WP_010595922.1,香茅醇假单胞菌(Pseudomonas citronellolis)P3B5的蛋白WP_061563290.1,恶臭假单胞菌(P. putida)B6-2的蛋白WP_008027336.1,恶臭假单胞菌(P. putida)F1的蛋白ABQ79984.1,恶臭假单胞菌(P. putida)GB-1的蛋白WP_012271104.1,恶臭假单胞菌(P. putida)SJTE-1的蛋白WP_014754112.1/ANI02794.1/ANI04816.1,睾丸酮单胞菌(C. testosteroni)P19的蛋白WP_057091786.1,鞘氨醇单胞菌(Sphingomonas sp.)KC8的蛋白WP_010128053.1。横线表示α螺旋,箭头表示β折叠;B:以4iqg.1.A为模型,以SWISSMODEL对蛋白SDR-X1进行同源建模获得的蛋白三维模型;C:蛋白SDR-X1的进化分析
Fig. 1 Multiple sequence alignment,advanced structural model and evolutionary analysis of SDR-X1 protein A:Multiple sequence alignment of short chain dehydrogenase from different microorganisms. Protein SDR-X1(WP_043267487.1)from P. citronellol SJTE-3,protein CAD85559.1 from N. europaea ATCC19718,protein WP_010595991.1 and WP_010595922.1 from R. ruber P14,protein WP_061563290.1 from P. citronellolis P3B5,protein WP_008027336.1 from P. putida B6-2,protein ABQ79984.1 from Pseudomonas putida F1,protein WP_012271104.1 from Pseudomonas putida GB-1,protein WP_014754112.1/ANI02794.1/ANI04816.1 from P. putida SJTE-1,protein WP_057091786.1 from C.testosteroni P19,and protein WP_010128053.1 from Sphingomonas KC8. The spiral marks the α-helix and the arrow marks the β-sheet. B:Having 4iqg.1.A as a model,and a three-dimensional model of protein obtained from homologous modeling of protein SDR-X1 with SWISSMODEL. C:Evolutionary analysis of the protein SDR-X1
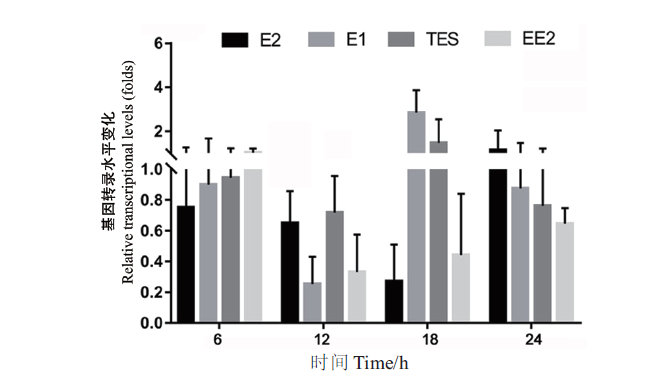
图2 基因sdr-x1在菌株SJTE-3以不同激素为碳源时的转录水平 碳源为10 μg/mL的激素(E1、E2、EE2或TES)或0.1%乙醇;将基因sdr-x1在乙醇为碳源培养基中的转录水平设为1.0。荧光定量PCR扩增结果采用2-∆∆Ct法处理数据。每组实验设3个平行,重复3次,计算平均值和标准误差
Fig. 2 Transcription levels of the gene sdr-x1 in strain SJTE-3 with different steroid hormones as carbon sources The carbon source is 10 μg/mL of steroid hormones(E1,E2,EE2 or TES)or 0.1% ethanol,and the transcription level of the gene sdr-x1 in ethanol as carbon source medium is set as 1.0. The results of fluorescence quantitative PCR amplification were processed by 2-∆∆Ct method. Three parallel experiments were set for each group and repeated three times,and based on which the mean and standard error were calculated

图3 重组菌株BL21-SDR-X1对E2的转化与重组蛋白SDR-X1纯化及其对E2的转化反应 A:重组菌株BL21-SDR-X1转化E2的效能测定。HPLC检测菌株BL21-SDR-X1在3、6、12、18、24 h对10 μg/mL 17β-雌二醇的转化效能。对照组为菌株带质粒pET28a的BL21(DE3)菌株。每组实验设3个平行,重复3次,计算平均值和标准误差;B:电泳检测亲和纯化的重组蛋白SDR-X1。泳道1:Maker;泳道2:诱导前的细胞裂解液;泳道3:诱导后的细胞裂解液;泳道4:细胞破碎离心沉淀;泳道5:细胞破碎离心上清;泳道6-7:蛋白流出液;泳道8-9:蛋白洗杂液;泳道10:蛋白洗脱液;C:重组蛋白SDR-X1转化E2的动力学曲线。以E2为底物,测定蛋白SDR-X1对不同浓度(0.01、0.0125、0.015、0.02、0.025、0.03、0.05、0.075、0.1 mmol/L)底物的反应速率。每组实验设3个平行,重复3次,计算平均值和标准误差
Fig. 3 Efficacy determination of recombinant strain BL21-SDR-X1 for transforming E2,purification of recombinant protein SDR-X1 and its transformation reaction to E2 A:The efficacy determination of recombinant strain BL21-SDR-X1 for transforming E2. The conversion efficiency of strain BL21-SDR-X1 to 10 μg/mL 17β-estradiol at 3,6,12,18 and 24 h is detected by HPLC. The control group is BL21(DE3)strain with plasmid pET28a. Three parallel experiments are set in each group and repeated for three times,and based on it the mean and standard error are calculated. B:Electrophoresis detection of affinity-purified recombinant protein SDR-X1. Lane 1:protein maker;lane 2:cell lysis solution before induction;lane 3:cell lysis solution after induction;lane 4:centrifugal precipitation of the crushed cells;lane 5:centrifugal supernatant of the crushed cells;lane 6-7:the solution after column loading;lane 8-9:the impurities;lane 10:protein eluent. C:Kinetic curve of recombinant protein SDR-X1 transforming E2. Using E2 as substrate,the effect of protein SDR-X1 on different concentration(0.0100,0.0125,0.0150,0.0200,0.0250,0.0300 0,0.0500 0,0.075,and 0.1 mmol/L)substrate is determined. Three parallel experiments are set for each group and repeated three times,and the mean and standard error are calculated
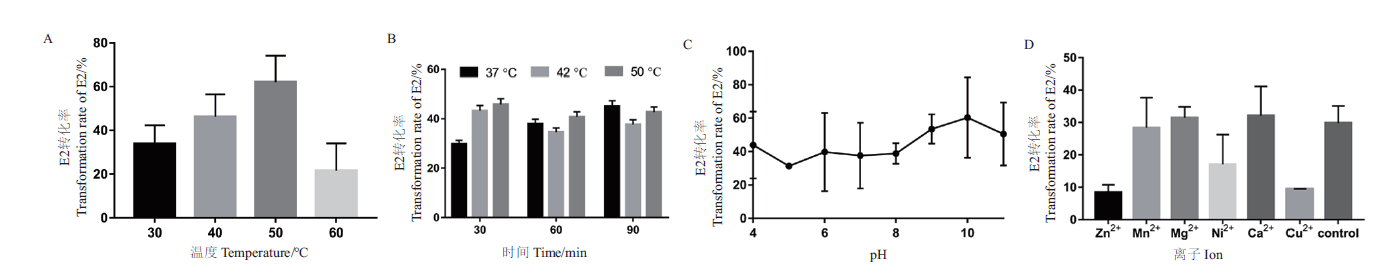
图4 重组酶SDR-X1转化E2的酶学性质测定 A:最适反应温度测定。不同温度(30℃,40℃,50℃,60℃)下,重组酶SDR-X1对E2的转化效率;B:温度耐受性测定。将重组酶SDR-X1在不同温度(30℃,42℃,50℃)下处理不同时间(30 min,60 min,90 min)后,其对E2的转化效率;C:最适反应pH测定。不同 pH(4.0-11.0)下,重组酶SDR-X1对E2的转化效率。D:二价金属离子影响测定。测定加入1 mmol/L不同金属离子(Zn2+,Mn2+,Mg2+,Ni2+,Ca2+,Cu2+),重组酶SDR-X1对E2的转化效率。每组实验设3个平行,重复3次,计算平均值和标准误差
Fig. 4 Determination of the enzymatic properties of E2 transformed by recombinant enzyme SDR-X1 A:Determination of optimal reaction temperature. The conversion efficiency of recombinase SDR-X1 at different temperature(30,40,50,and 60℃)was detected with E2 as substrate. B:Temperature tolerance was measured. The conversion efficiency of recombinase SDR-X1 treated with different temperature(37,42,and 50℃)and time(30,60,and 90 min)was detected with E2 as substrate. C:Optimal reaction pH determination. The conversion efficiency of recombinase SDR-X1 at different pH value(pH4-11)was detected with E2 as substrate. D:Determination of influence of divalent metal ions. The conversion efficiency of recombinase SDR-X1 under the action of different metal ion(Zn2+,Mn2+,Mg2+,Ni2+,Ca2+,and Cu2+)at 1 mmol/L was determined. Three experiments were set in parallel for each group and repeated for three times. The mean value and standard error are calculated
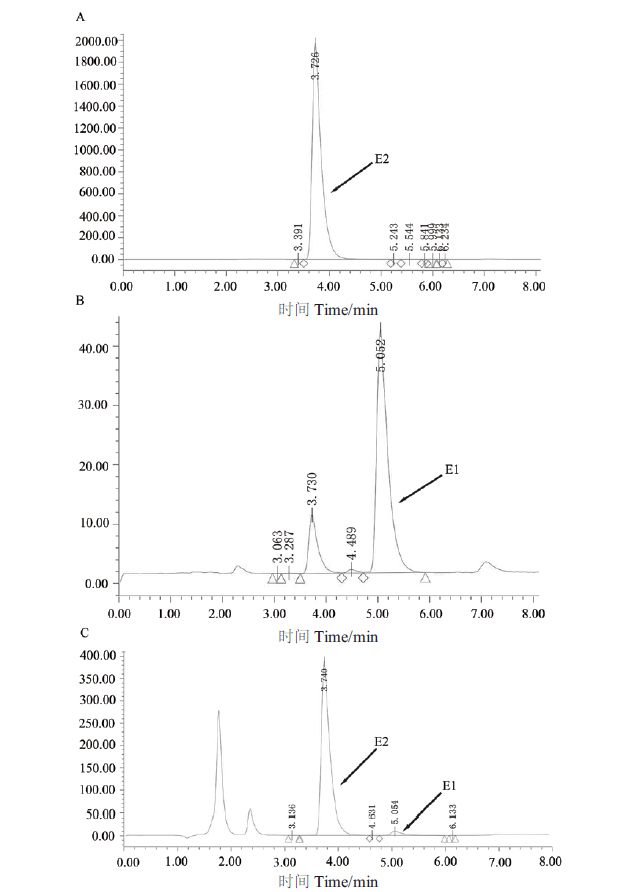
图5 HPLC分析重组酶SDR-X1转化E2的反应产物 A:HPLC检测50 μg/mL的E2 的色谱图;B:HPLC检测50 μg/mL的E1的色谱图;C:HPLC检测重组酶SDR-X1转化E2的反应产物。200 μL反应体系中含有 0.1 mmol/L E2、0.2 mmol/L NAD+和100 μg/mL蛋白SDR-X1,42℃反应15 min后加入1/3体积的乙腈,振荡混匀后取50 μL进行HPLC检测图谱
Fig. 5 HPLC analysis of reaction products of conversion of E2 by recombinase SDR-X1 A:The HPLC map of 50 μg/mL E2. B:The HPLC map of 50 μg/mL E1. C:The HPLC map of the products of E2 convered by recombinase SDR-X1. The reaction solution is 0.1 mmol/L E2,0.2 mmol/L NAD+ and 100 μg/mL protein SDR-X1 in a 200 μL reaction system at 42℃ for 15 min. Then adding 1/3 volume of acetonitrile to the reaction solution,swirling and mixing,50 μL sample is taken to detect degradation of E2 by HPLC
| [1] |
Wise A, O'Brien K, Woodruff T. Are oral contraceptives a significant contributor to the estrogenicity of drinking water?[J]. Environ Sci Technol, 2011, 45(1):51-60.
doi: 10.1021/es1014482 URL |
| [2] |
Kiyama R, Wada-Kiyama Y. Estrogenic endocrine disruptors:Molecular mechanisms of action[J]. Environ Int, 2015, 83:11-40.
doi: 10.1016/j.envint.2015.05.012 URL |
| [3] |
Chen YL, Wang CH, Yang FC, et al. Identification of Comamonas testosteroni as an androgen degrader in sewage[J]. Sci Rep, 2016, 6:35386.
doi: 10.1038/srep35386 URL |
| [4] |
Luine VN. Estradiol and cognitive function:past, present and future[J]. Horm Behav, 2014, 66(4):602-618.
doi: 10.1016/j.yhbeh.2014.08.011 URL |
| [5] |
Donova MV, Egorova OV. Microbial steroid transformations:current state and prospects[J]. Appl Microbiol Biotechnol, 2012, 94(6):1423-1447.
doi: 10.1007/s00253-012-4078-0 URL |
| [6] | Chen YL, Fu HY, Lee TH, et al. Estrogen degraders and estrogen degradation pathway identified in an activated sludge[J]. Appl Environ Microbiol, 2018, 84(10):e00001-e00018. |
| [7] |
Bai X, Acharya K. Removal of seven endocrine disrupting chemicals(EDCs)from municipal wastewater effluents by a freshwater green alga[J]. Environ Pollut, 2019, 247:534-540.
doi: 10.1016/j.envpol.2019.01.075 URL |
| [8] |
Xiong W, Yin C, Wang Y, et al. Characterization of an efficient estrogen-degrading bacterium Stenotrophomonas maltophilia SJTH1 in saline-, alkaline-, heavy metal-contained environments or solid soil and identification of four 17β-estradiol-oxidizing dehydrogenases[J]. J Hazard Mater, 2020, 385:121616.
doi: 10.1016/j.jhazmat.2019.121616 URL |
| [9] |
Wang P, Zheng D, Liang R. Isolation and characterization of an estrogen-degrading Pseudomonas putida strain SJTE-1[J]. 3 Biotech, 2019, 9(2):61.
doi: 10.1007/s13205-018-1537-z URL |
| [10] | Li SY, Liu J, Sun MX, et al. Isolation, characterization, and degradation performance of the 17β-estradiol-degrading bacterium Novosphingobium sp. E2S[J]. Int J Environ Res Public Heal, 2017, 14(2):115. |
| [11] |
Xiong WL, Yin C, Peng WL, et al. Characterization of an 17β-estradiol-degrading bacterium Stenotrophomonas maltophilia SJTL3 tolerant to adverse environmental factors[J]. Appl Microbiol Biotechnol, 2020, 104(3):1291-1305.
doi: 10.1007/s00253-019-10281-8 URL |
| [12] |
Mascotti ML, Palazzolo MA, Bisogno FR, et al. Biotransformation of dehydro-epi-androsterone by Aspergillus parasiticus:Metabolic evidences of BVMO activity[J]. Steroids, 2016, 109:44-49.
doi: 10.1016/j.steroids.2016.03.018 pmid: 27025973 |
| [13] | Liu WJ, Chen Q, He N, et al. Removal and biodegradation of 17β-estradiol and diethylstilbestrol by the freshwater microalgae Raphidocelis subcapitata[J]. Int J Environ Res Public Heal, 2018, 15(3):452. |
| [14] |
Wang PH, Yu CP, Lee TH, et al. Anoxic androgen degradation by the denitrifying bacterium Sterolibacterium denitrificans via the 2, 3-seco pathway[J]. Appl Environ Microbiol, 2014, 80(11):3442-3452.
doi: 10.1128/AEM.03880-13 URL |
| [15] |
Wang PP, Zheng DN, Peng WL, et al. Characterization of 17β-hydroxysteroid dehydrogenase and regulators involved in estrogen degradation in Pseudomonas putida SJTE-1[J]. Appl Microbiol Biotechnol, 2019, 103(5):2413-2425.
doi: 10.1007/s00253-018-9543-y URL |
| [16] |
Ye X, Wang H, Kan J, et al. A novel 17β-hydroxysteroid dehydrogenase in Rhodococcus sp. P14 for transforming 17β-estradiol to estrone[J]. Chem Biol Interact, 2017, 276:105-112.
doi: 10.1016/j.cbi.2017.06.010 URL |
| [17] | Liu C, Liu K, Zhao C, et al. The characterization of a short chain dehydrogenase/reductase(SDRx)in Comamonas testosteroni[J]. Toxicol Rep, 2020, 7:460-467. |
| [18] |
Yu YH, Liu CZ, Wang BX, et al. Characterization of 3, 17β-hydroxysteroid dehydrogenase in Comamonas testosteroni[J]. Chem Biol Interact, 2015, 234:221-228.
doi: 10.1016/j.cbi.2015.01.005 URL |
| [19] |
Pan T, Huang P, Xiong G, et al. Isolation and identification of a repressor TetR for 3, 17β-HSD expressional regulation in Comamonas testosteroni[J]. Chem Biol Interact, 2015, 234:205-212.
doi: 10.1016/j.cbi.2014.12.034 URL |
| [20] |
Kavanagh KL, Jörnvall H, Persson B, et al. Medium- and short-chain dehydrogenase/reductase gene and protein families[J]. Cell Mol Life Sci, 2008, 65(24):3895-3906.
doi: 10.1007/s00018-008-8588-y pmid: 19011750 |
| [21] |
Gräff M, Buchholz PCF, Stockinger P, et al. The Short-chain Dehydrogenase/Reductase Engineering Database(SDRED):a classification and analysis system for a highly diverse enzyme family[J]. Proteins, 2019, 87(6):443-451.
doi: 10.1002/prot.v87.6 URL |
| [22] | Zheng DN, Wang XL, Wang PP, et al. Genome sequence of Pseudomonas citronellolis SJTE-3, an estrogen-and polycyclic aromatic hydrocarbon-degrading bacterium[J]. Genome Announc, 2016, 4(6):e01373-16. |
| [23] |
Hall BG. Building phylogenetic trees from molecular data with MEGA[J]. Mol Biol Evol, 2013, 30(5):1229-1235.
doi: 10.1093/molbev/mst012 URL |
| [24] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J]. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [25] |
Xu J, Zhang L, Hou J, et al. iTRAQ-based quantitative proteomic analysis of the global response to 17β-estradiol in estrogen-degradation strain Pseudomonas putida SJTE-1[J]. Sci Rep, 2017, 7:41682.
doi: 10.1038/srep41682 URL |
| [26] |
Kallberg Y, Oppermann U, Persson B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models[J]. Febs J, 2010, 277(10):2375-2386.
doi: 10.1111/j.1742-4658.2010.07656.x pmid: 20423462 |
| [27] | Mythen SM, Devendran S, Méndez-García C, et al. Targeted synjournal and characterization of a gene cluster encoding NAD(P)H-dependent 3α-, 3β-, and 12α-hydroxysteroid dehydrogenases from Eggerthella CAG:298, a gut metagenomic sequence[J]. Appl Environ Microbiol, 2018, 84(7):e02475-17. |
| [28] |
Zhao BH, Sun Q, Chen J, et al. 17 beta-estradiol biodegradation by anaerobic granular sludge:Effect of iron sources[J]. Sci Rep, 2020, 10:7777.
doi: 10.1038/s41598-020-64557-5 URL |
| [1] | 马青云, 江旭, 李情情, 宋金龙, 周义清, 阮志勇. 烟嘧磺隆降解菌Chryseobacterium sp. LAM-M5的分离、鉴定及其降解机理研究[J]. 生物技术通报, 2022, 38(2): 113-122. |
| [2] | 牛鸿宇, 舒倩, 杨海君, 颜智勇, 谭菊. 一株十二烷基硫酸钠高效降解菌的分离鉴定、降解特性及代谢途径研究[J]. 生物技术通报, 2022, 38(12): 287-299. |
| [3] | 肖小双, 安雪姣, 叶晗媛, 王林平, 钟斌, 张庆华. 废水中硫氰酸盐的微生物降解研究进展[J]. 生物技术通报, 2021, 37(2): 224-235. |
| [4] | 王豪, 唐禄鑫, 马鸿飞, 钱坤, 司静, 崔宝凯. 东方栓孔菌漆酶的固定化及其对不同类型染料的脱色作用[J]. 生物技术通报, 2021, 37(11): 142-157. |
| [5] | 吴敏, 唐洁, 胡琼, 雷丹, 张庆. 表面活性剂对琼式不动杆菌LH-1-1降解溴氰菊酯的影响[J]. 生物技术通报, 2021, 37(1): 215-222. |
| [6] | 岳丽晓, 李登云, 张晶晶, 仝雷. 一株敌草隆降解菌的分离及其应用潜能探索[J]. 生物技术通报, 2020, 36(6): 110-119. |
| [7] | 陈锐, 瞿佳, 孙晓宇, 邓媛, 门欣, 赵玲侠, 沈卫荣. 氯氰菊酯降解菌草酸青霉SSCL-5分离鉴定及降解特性[J]. 生物技术通报, 2020, 36(6): 120-127. |
| [8] | 高超, 郝孔利, 赵宇婷, 毛樱翔, 池明眼, 张杰. 一株能够降解聚乙烯的霍氏肠杆菌的鉴定及分析[J]. 生物技术通报, 2020, 36(10): 99-104. |
| [9] | 王亚妮, 宋金龙, 韩刚, 穆迎春, 江旭, 王金耀, 阮志勇, 李乐. 孔雀石绿降解菌群多样性及高效降解菌的降解特性分析[J]. 生物技术通报, 2019, 35(9): 150-155. |
| [10] | 刘娜, 刘志敏, 宋东辉. 石油烃降解菌对邻苯二酚、苯甲酸钠降解特性的研究[J]. 生物技术通报, 2019, 35(9): 156-164. |
| [11] | 任磊, 刘斌, 蔺中, 甄珍, 刘月廉, 胡汉桥, 闫艳春. 一株耐盐对硝基苯酚降解菌的分离及其降解机理研究[J]. 生物技术通报, 2019, 35(9): 184-193. |
| [12] | 郭亚男, 张馨予, 胥梦, 王继华. 低温萘降解菌的筛选、鉴定及降解条件优化[J]. 生物技术通报, 2019, 35(7): 100-107. |
| [13] | 刘亚兰, 段梦洁, 林晓珊, 张毅. 聚乙烯醇降解细菌筛选及其降解特性[J]. 生物技术通报, 2019, 35(6): 91-98. |
| [14] | 张珊珊, 姚小龙, 王珂, 尤雅. 1株杀鲑气单胞菌杀鲑亚种对甲酸乙酯的降解研究[J]. 生物技术通报, 2019, 35(5): 109-117. |
| [15] | 田晶, 徐小琳, 康彦顺, 汤伟华, 刘思琪. 广谱性多环芳烃降解真菌Aspergillus flavus AD-X-1的筛选及其性能研究[J]. 生物技术通报, 2018, 34(8): 115-122. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||