








生物技术通报 ›› 2023, Vol. 39 ›› Issue (10): 197-208.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0364
王兵1( ), 赵会纳1, 余婧1, 陈杰1, 骆梅2, 雷波1(
), 赵会纳1, 余婧1, 陈杰1, 骆梅2, 雷波1( )
)
收稿日期:2023-04-19
出版日期:2023-10-26
发布日期:2023-11-28
通讯作者:
雷波,女,博士,研究员,研究方向:烟草分子生物学;E-mail: leibo_1981@163.com作者简介:王兵,男,博士,助理研究员,研究方向:烟草分子生物学;E-mail: vipwb1519599@163.com
基金资助:
WANG Bing1( ), ZHAO Hui-na1, YU Jing1, CHEN Jie1, LUO Mei2, LEI Bo1(
), ZHAO Hui-na1, YU Jing1, CHEN Jie1, LUO Mei2, LEI Bo1( )
)
Received:2023-04-19
Published:2023-10-26
Online:2023-11-28
摘要:
作物叶芽受到分生组织调控,调控叶芽是作物增产的有效措施之一。目前关于烟草分生组织调控的分子机理研究偏少,可用于株型改良的种质资源缺乏。本研究通过CRISPR/Cas9编辑系统靶向突变烟草REVOLUTA(REV)基因,分别构建两个不同REV单靶点序列C15NtREV和C16NtREV,通过农杆菌介导的叶盘转化方法获得再生苗,利用PCR测序鉴定转基因阳性单株,测序结果表明Ko-C15Ntrev突变体在NtREV氨基酸第26位置之后发生移码突变,而Ko-C16Ntrev突变体在NtREV氨基酸第60位置之后发生移码突变。此外,借助扫描电镜分别观测两个单靶点纯合突变体顶芽表型,结果表明烟草Ko-C15Ntrev双拷贝同源突变体出现顶芽缺失和叶片畸形的表型,而Ko-C16Ntrev单拷贝同源突变体未表现出顶芽缺失,但顶芽发育迟缓。相较于野生型烟草,Ko-C16Ntrev突变体自然株高较野生型增加3.76%,Ko-C16Ntrev突变体叶片数和腋芽鲜重分别较野生型减少21.47%和23.41%,且均达到极显著差异,说明NtREV参与烟草顶端分生组织发育,进而调节叶和腋芽发育。这些突变体为后续研究烟草的叶芽发育分子机理提供了重要研究材料。
王兵, 赵会纳, 余婧, 陈杰, 骆梅, 雷波. 利用CRISPR/Cas9系统研究REVOLUTA参与烟草叶芽发育的调控[J]. 生物技术通报, 2023, 39(10): 197-208.
WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System[J]. Biotechnology Bulletin, 2023, 39(10): 197-208.
| 引物Primer | 序列Sequence(5'-3') | 作用Purpose |
|---|---|---|
| C16NtREV-T1F | ATTGGCTCGATATTCGACAGAATAGGG | 靶点接头引物 NtREV target construction |
| C16NtREV-T1R | AAACCCCTATTCTGTCGAATATCGAGC | |
| C15NtREV-T1F | ATTGACAGTAGTGGAAAGTATGTCCGG | |
| C15NtREV-T1R | AAACCCGGACATACTTTCCACTACTGT | |
| 1300-gRNA-F | CCAGTCACGACGTTGTAAAAC | SgRNA表达盒检测 Detection of SgRNA expression kit |
| 1300-gRNA-R | CAATGAATTTCCCATCGTCGAG | |
| Hyg-F | CGATTGCGTCGCATCGACC | 潮霉素抗性基因检测 Detection of hygromycin resistance gene |
| Hyg-R | TTCTACAACCGGTCGCGGAG | |
| C16NtREV-CRISPR test-F | TTTTGGTTTGGGATTTTGAGG | 检测转基因突变 Detection of transgenic mutations |
| C16NtREV-CRISPR test-R | CATTGTTCAATTTGGATTACTCC | |
| C16NtREV-CRISPR test-R | GGAGCAATCAAAATTGAACAGCG | |
| C15NtREV-CRISPR test-F | CTATGGTTGCACAGCAGCACA | |
| C15NtREV-CRISPR test-R | ACTTCATCCATCACTGATCTAAC | |
| C15NtREV-CRISPR test-R | ACTAGAAGCACAGTGATATATTC |
表1 本研究引物序列及用途
Table 1 Sequences and purpose of primers in this study
| 引物Primer | 序列Sequence(5'-3') | 作用Purpose |
|---|---|---|
| C16NtREV-T1F | ATTGGCTCGATATTCGACAGAATAGGG | 靶点接头引物 NtREV target construction |
| C16NtREV-T1R | AAACCCCTATTCTGTCGAATATCGAGC | |
| C15NtREV-T1F | ATTGACAGTAGTGGAAAGTATGTCCGG | |
| C15NtREV-T1R | AAACCCGGACATACTTTCCACTACTGT | |
| 1300-gRNA-F | CCAGTCACGACGTTGTAAAAC | SgRNA表达盒检测 Detection of SgRNA expression kit |
| 1300-gRNA-R | CAATGAATTTCCCATCGTCGAG | |
| Hyg-F | CGATTGCGTCGCATCGACC | 潮霉素抗性基因检测 Detection of hygromycin resistance gene |
| Hyg-R | TTCTACAACCGGTCGCGGAG | |
| C16NtREV-CRISPR test-F | TTTTGGTTTGGGATTTTGAGG | 检测转基因突变 Detection of transgenic mutations |
| C16NtREV-CRISPR test-R | CATTGTTCAATTTGGATTACTCC | |
| C16NtREV-CRISPR test-R | GGAGCAATCAAAATTGAACAGCG | |
| C15NtREV-CRISPR test-F | CTATGGTTGCACAGCAGCACA | |
| C15NtREV-CRISPR test-R | ACTTCATCCATCACTGATCTAAC | |
| C15NtREV-CRISPR test-R | ACTAGAAGCACAGTGATATATTC |
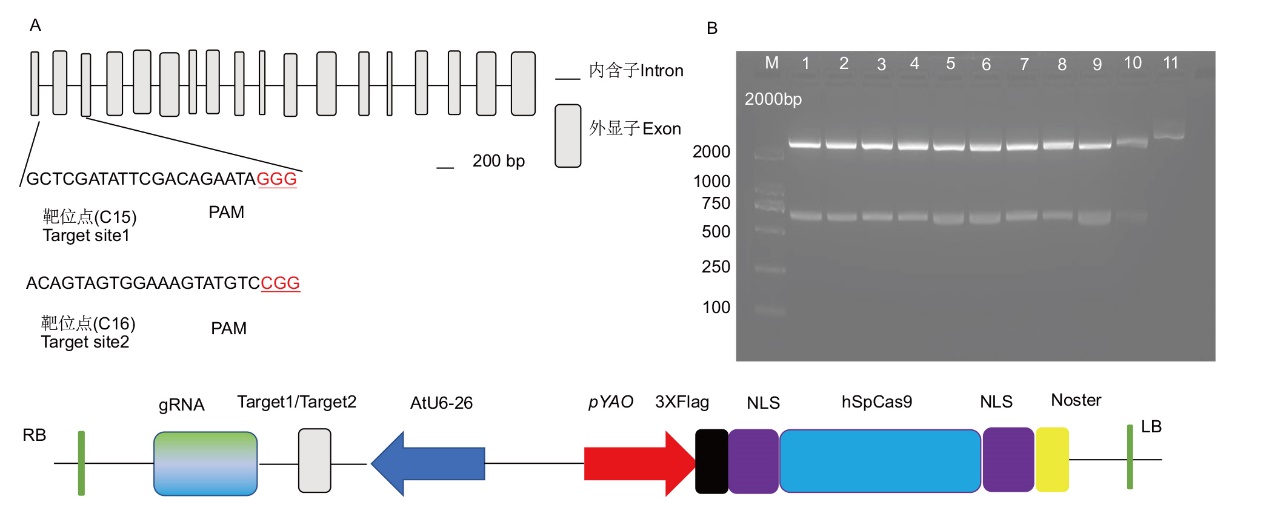
图2 烟草REV结构示意图及靶位点选择与载体构建 A:REV基因外显子和内含子示意图以及靶点编辑序列所在位置;B :REV基因靶点编辑序列的双酶切核酸电泳
Fig. 2 Schematic structure and target site selection of NtREV and vector construction A: Schematic diagram of REV gene exons and introns, as well as the locations of the target editing sequences. B: Nucleic acid electrophoresis of double enzyme digestion of REV gene the target editing sequences
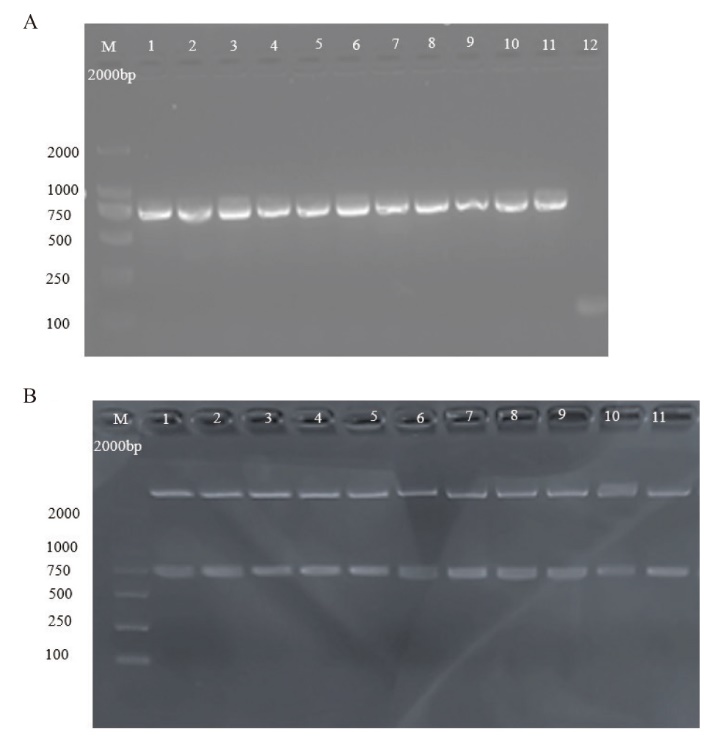
图3 植物双元表达载体pCAMBIA1300-pYAO-Cas9-NtREV的构建 A:pCAMBIA1300-pYAO-Cas9-NtREV的PCR 扩增的核酸电泳; B :pCAMB-IA1300-pYAO-Cas9-NtREV双酶切的核酸电泳
Fig. 3 Construction of the plant binary expression vector pCAMBIA1300-pYAO-Cas9-NtREV A: Nucleic acid electrophoresis of PCR amplification of pCAMBIA1300-pYAO-Cas9-NtREV. B: Nucleic acid electrophoresis of double enzyme digestion of pCAMBIA1300-pYAO-Cas9-NtREV
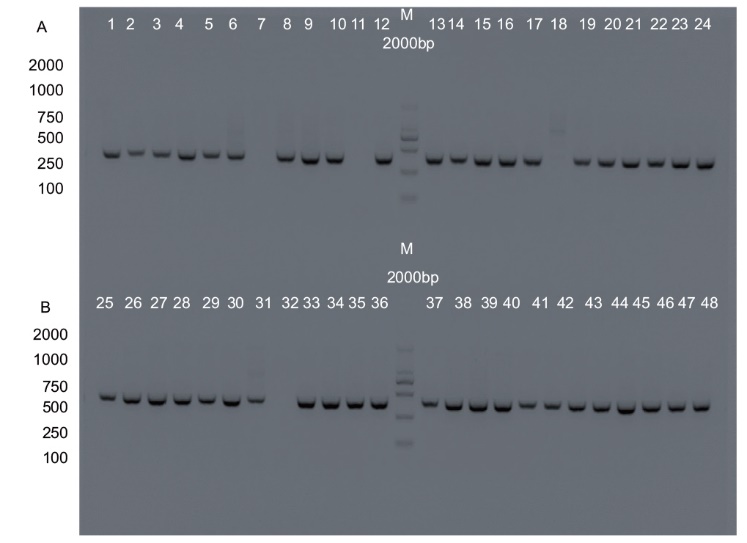
图4 转基因植株检测的核酸电泳 A :REV基因靶点C15编辑植株检测的核酸电泳;B :REV基因靶点C16编辑植株检测的核酸电泳
Fig. 4 Nucleic acid electrophoresis for the detection of transgenic plants A: Nucleic acid electrophoresis for detecting plant editing of the REV gene at target C15. B: Nucleic acid electrophoresis for detecting plant editing of the REV gene at target C16

图5 CRISPR/Cas9编辑NtREV基因突变体形式分析 A :REV基因靶点C16序列的突变情况 ;B :REV基因靶点C15序列的突变情况。下划线表示PAM序列,蓝色字表示替换碱基,红色字表示插入碱基,-表示缺失的碱基
Fig. 5 Mutation form analysis of NtREV edited by the CRISPR/Cas9 A: Mutation of REV target C16 sequence; B: mutation of REV target C15 sequence. Underscores indicate PAM sequences, blue words indicate replacement bases, red words indicate insertion bases, and - indicates missing bases

图6 突变体材料和野生型材料的叶形态表型A-F:双拷贝同源突变体材料叶发育形态,包括漏斗形叶、荷叶形叶等形态;G-H:单拷贝同源突变体材料叶发育形态; I:对应时期野生型生长状态;标尺:1 cm
Fig. 6 Leaf morphological phenotypes of mutant and wild-type materials A-F: Leaf development morphology of the double-copy homologous mutants, including funnel-shaped leaves, lotus-shaped leaves and other morphologies; G-H: leaf development morphology of the single-copy homologous mutants; I: wild type growth state in the corresponding period; bar=1 cm
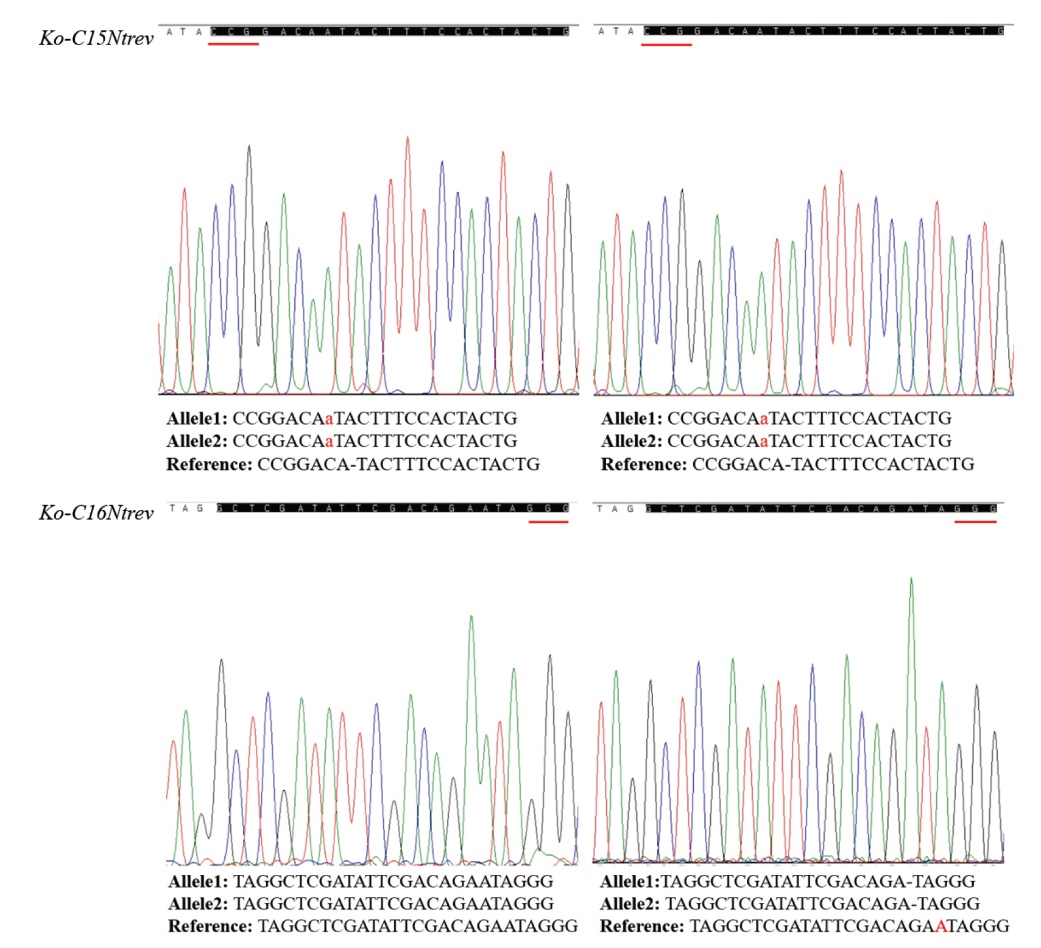
图7 烟草Ko-C15Ntrev和Ko-C16Ntrev突变体编辑形式Ko-C15Ntrev编辑形式为双拷贝同源突变体;Ko-C16Ntrev编辑形式为单拷贝同源突变体
Fig. 7 Editing forms of Ko-C15Ntrev and Ko-C16Ntrev mutants in tobacco The double-copy homologous mutants of Ko-C15NtREV; Ko-C16NtREV editing form is the single-copy homologous mutants
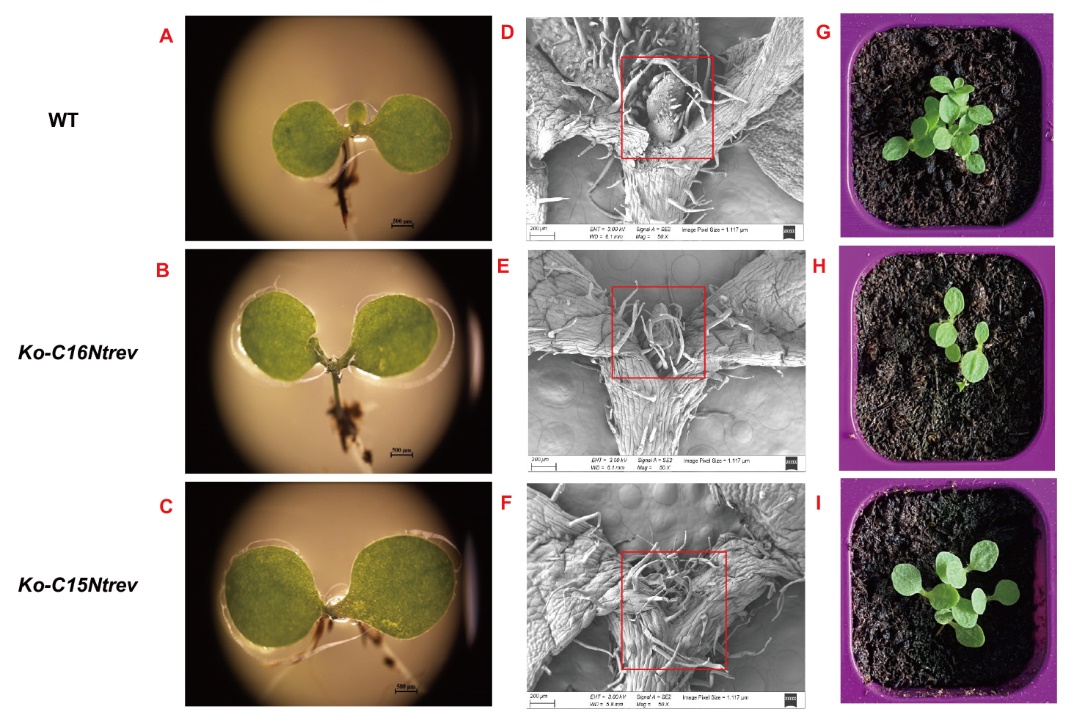
图8 突变体材料和野生型材料的顶芽观测 A,B,C分别为体式显微镜下烟草苗期野生型WT、Ko-C16Ntrev和Ko-C15Ntrev突变体顶芽表型;D,E,F分别为扫描电子显微镜下烟草苗期WT、Ko-C16Ntrev和Ko-C15Ntrev突变体顶端分生组织;G,H,I分别为上述表型对应的烟草苗期WT、Ko-C16Ntrev和Ko-C15Ntrev突变体材料
Fig. 8 Observations of apical buds of mutant and wild-type ones A, B, C: Apical bud phenotypes of tobacco seedling wild-type, Ko-C16Ntrev and Ko-C15Ntrev mutant under stereomicroscope, respectively; D, E, F: apical meristem phenotypes of tobacco seedling wild-type,Ko-C16Ntrev and Ko-C15Ntrev mutant under scanning electron microscopy, respectively; G, H, I: tobacco seedling wild-type, Ko-C16Ntrev and Ko-C15Ntrev mutant corresponding to the above phenotypes, respectively

图9 Ko-C16Ntrev突变体的自然株高、叶数、腋芽鲜重表型 A,B,C分别为WT和Ko-C16Ntrev突变体的自然株高、叶数、腋芽鲜重的统计;D,E分别为WT和Ko-C16Ntrev突变体侧芽表型图。数据表示平均值 ± SD,其中 n ≥ 10 ;**表示显著性 P<0.01;标尺:5 cm
Fig. 9 Phenotypes of plant height, leaf numbers and fresh weight of C16Ntrev mutants A, B, C: Statistical analysis of plant height, leaf numbers and fresh weight content of axillary buds of WT and Ko-C16Ntrev mutants. D and E: The phenotype of lateral buds of WT and Ko-C16Ntrev mutants, respectively. Bar= 5 cm. The data represent mean ± SD, n ≥ 10; ** indicates significance P<0.01
| [1] |
Teichmann T, Muhr M. Shaping plant architecture[J]. Front Plant Sci, 2015, 6: 233.
doi: 10.3389/fpls.2015.00233 pmid: 25914710 |
| [2] |
王兵, 赵会纳, 余婧, 等. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1112 |
|
Wang B, Zhao HN, Yu J, et al. Research progress in the regulation of plant branch development[J]. Biotechnol Bull, 2023, 39(5): 14-22.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1112 |
|
| [3] |
Luo M, Gilbert B, Ayliffe M. Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants[J]. Plant Cell Rep, 2016, 35(7): 1439-1450.
doi: 10.1007/s00299-016-1989-8 pmid: 27146973 |
| [4] | 刘耀光, 李构思, 张雅玲, 等. CRISPR/Cas植物基因组编辑技术研究进展[J]. 华南农业大学学报, 2019, 40(5): 38-49. |
| Liu YG, Li GS, Zhang YL, et al. Current advances on CRISPR/Cas genome editing technologies in plants[J]. J South China Agric Univ, 2019, 40(5): 38-49. | |
| [5] | 杨雪, 孙雅佩, 王政博, 等. CRISPR/Cas9介导靶向敲除拟南芥REVOLUTA基因突变体的鉴定[J]. 分子植物育种, 2021, 19(3): 867-873. |
| Yang X, Sun YP, Wang ZB, et al. Identification of knockout of REVOLUTA mutant caused by CRISPR/Cas9 in Arabidopsis[J]. Mol Plant Breed, 2021, 19(3): 867-873. | |
| [6] | Amoo O, 胡利民, 翟云孤, 等. 利用基因编辑技术研究BRANCHED1参与油菜分枝过程的调控[J]. 生物技术通报, 2022, 38(4): 97-105. |
| Amoo O, Hu LM, Zhai YG, et al. Regulation of shoot branching by BRANCHED1 in Brassica napus based on gene editing technology[J]. Biotechnol Bull, 2022, 38(4): 97-105. | |
| [7] |
Prigge MJ, Otsuga D, Alonso JM, et al. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development[J]. Plant Cell, 2005, 17(1): 61-76.
doi: 10.1105/tpc.104.026161 URL |
| [8] |
Zhong RQ, Ye ZH. Regulation of HD-ZIP III genes by microRNA 165[J]. Plant Signal Behav, 2007, 2(5): 351-353.
doi: 10.4161/psb.2.5.4119 URL |
| [9] |
Kim J, Jung JH, Reyes JL, et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems[J]. Plant J, 2005, 42(1): 84-94.
doi: 10.1111/tpj.2005.42.issue-1 URL |
| [10] | 杜玉梁, 陈叶, 刘彩虹, 等. 植物维管组织形态建成的分子调控机制[J]. 北京林业大学学报, 2014, 36(3): 142-150. |
| Du YL, Chen Y, Liu CH, et al. Molecular regulation mechanism of vascular pattern formation in plant[J]. J Beijing For Univ, 2014, 36(3): 142-150. | |
| [11] |
Talbert PB, Adler HT, Parks DW, et al. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana[J]. Development, 1995, 121(9): 2723-2735.
doi: 10.1242/dev.121.9.2723 pmid: 7555701 |
| [12] |
Otsuga D, DeGuzman B, Prigge MJ, et al. REVOLUTA regulates meristem initiation at lateral positions[J]. Plant J, 2001, 25(2): 223-236.
doi: 10.1046/j.1365-313x.2001.00959.x pmid: 11169198 |
| [13] |
Williams L, Grigg SP, Xie MT, et al. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes[J]. Development, 2005, 132(16): 3657-3668.
doi: 10.1242/dev.01942 pmid: 16033795 |
| [14] |
Zhong RQ, Ye ZH. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels[J]. Plant Cell Physiol, 2004, 45(4): 369-385.
doi: 10.1093/pcp/pch051 URL |
| [15] |
Hawker NP, Bowman JL. Roles for Class III HD-Zip and KANADI genes in Arabidopsis root development[J]. Plant Physiol, 2004, 135(4): 2261-2270.
doi: 10.1104/pp.104.040196 URL |
| [16] |
Shi BH, Zhang C, Tian CH, et al. Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis[J]. PLoS Genet, 2016, 12(7): e1006168.
doi: 10.1371/journal.pgen.1006168 URL |
| [17] | Zhang C, Wang J, Wenkel S, et al. Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators[J]. Development, 2018, 145(24): dev158352. |
| [18] |
Zhang C, Fan LS, Le BH, et al. Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis[J]. Dev Cell, 2020, 55(5): 603-616.e5.
doi: 10.1016/j.devcel.2020.10.019 pmid: 33232670 |
| [19] | 陈浣, 吕婧, 孙玉合. 普通烟草HD-Zip III家族全基因组鉴定和表达分析[J]. 基因组学与应用生物学, 2017, 36(8): 3034-3041. |
| Chen H, Lyu J, Sun YH. Genome-wide identification and expression analysis of the HD-zip III gene family in Nicotiana tobacum[J]. Genom Appl Biol, 2017, 36(8): 3034-3041. | |
| [20] |
Ma XL, Zhang QY, Zhu QL, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants[J]. Mol Plant, 2015, 8(8): 1274-1284.
doi: 10.1016/j.molp.2015.04.007 pmid: 25917172 |
| [21] | 王兵, 程子义, 张蕾, 等. 过表达毛白杨线粒体APX基因烟草提高抗逆能力的研究[J]. 北京林业大学学报, 2020, 42(7): 33-39. |
| Wang B, Cheng ZY, Zhang L, et al. Tobacco overexpression Populus tomentosa mitochondria ascorbate peroxidase improving stress resistance[J]. J Beijing For Univ, 2020, 42(7): 33-39. | |
| [22] | 王兵.毛白杨天冬氨酸蛋白酶(PtoAED3)调控次生木质部发育机制研究[D]. 北京: 北京林业大学, 2020. |
| Wang B.Study on the mechanism of secondary xylem development regulated by aspartic protease(PtoAED3) of Populus tomentosa[D]. Beijing: Beijing Forestry University, 2020. | |
| [23] |
Zhang T, You J, Zhang Y, et al. LF1 regulates the lateral organs polarity development in rice[J]. New Phytol, 2021, 231(3): 1265-1277.
doi: 10.1111/nph.17220 pmid: 33469925 |
| [24] |
张晨, 雷展, 李凯, 等. CRISPR/Cas9系统中的脱靶效应及检测技术研究进展[J]. 生物技术通报, 2020, 36(3): 78-87.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0871 |
|
Zhang C, Lei Z, Li K, et al. Research progress on off-target effects and detection techniques in CRISPR/Cas9 systems[J]. Biotechnol Bull, 2020, 36(3): 78-87.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0871 |
|
| [25] |
Mukherjee K, Bürglin TR. MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins[J]. Plant Physiol, 2006, 140(4): 1142-1150.
doi: 10.1104/pp.105.073833 pmid: 16607028 |
| [26] |
Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains[J]. Structure, 2009, 17(10): 1282-1294.
doi: 10.1016/j.str.2009.08.011 pmid: 19836329 |
| [27] |
McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation[J]. Annu Rev Physiol, 2010, 72: 625-645.
doi: 10.1146/annurev-physiol-021909-135922 pmid: 20148691 |
| [28] |
Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins[J]. Science, 2009, 324(5930): 1068-1071.
doi: 10.1126/science.1173041 URL |
| [29] |
Brandt R, Salla-Martret M, Bou-Torrent J, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses[J]. Plant J, 2012, 72(1): 31-42.
doi: 10.1111/tpj.2012.72.issue-1 URL |
| [1] | 王宝宝, 王海洋. 理想株型塑造之于玉米耐密改良[J]. 生物技术通报, 2023, 39(8): 11-30. |
| [2] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [3] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| [4] | 石佳鑫, 刘凯, 朱金洁, 祁显涛, 谢传晓, 刘昌林. 基因编辑技术改良玉米株型增加杂交种产量[J]. 生物技术通报, 2023, 39(8): 62-69. |
| [5] | 施炜涛, 姚春鹏, 魏文康, 王蕾, 房元杰, 仝钰洁, 马晓姣, 蒋文, 张晓爱, 邵伟. 利用CRISPR/Cas9技术构建MDH2敲除细胞株及抗呕吐毒素效应研究[J]. 生物技术通报, 2023, 39(7): 307-315. |
| [6] | 刘晓燕, 祝振亮, 史广宇, 华梓宇, 杨晨, 张涌, 刘军. 乳腺生物反应器的表达优化策略[J]. 生物技术通报, 2023, 39(5): 77-91. |
| [7] | 程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291. |
| [8] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [9] | 李双喜, 华进联. 抗猪繁殖与呼吸障碍综合征基因编辑猪研究进展[J]. 生物技术通报, 2023, 39(10): 50-57. |
| [10] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [11] | 刘静静, 刘晓蕊, 李琳, 王盈, 杨海元, 戴一凡. 利用CRISPR/Cas9技术建立OXTR基因敲除猪胎儿成纤维细胞系[J]. 生物技术通报, 2022, 38(6): 272-278. |
| [12] | Olalekan Amoo, 胡利民, 翟云孤, 范楚川, 周永明. 利用基因编辑技术研究BRANCHED1参与油菜分枝过程的调控[J]. 生物技术通报, 2022, 38(4): 97-105. |
| [13] | 丁亚群, 丁宁, 谢深民, 黄梦娜, 张昱, 张勤, 姜力. Vps28基因敲除小鼠模型的构建及其对泌乳和免疫性状影响的研究[J]. 生物技术通报, 2022, 38(3): 164-172. |
| [14] | 燕炯, 冯晨毅, 高学坤, 许祥, 杨佳敏, 陈朝阳. 基于CRISPR/Cas9技术构建Plin1基因敲除小鼠模型及表型分析[J]. 生物技术通报, 2022, 38(3): 173-180. |
| [15] | 钟菁, 孙玲玲, 张姝, 蒙园, 支怡飞, 涂黎晴, 徐天鹏, 濮黎萍, 陆阳清. 应用CRISPR/Cas9技术敲除Mda5基因对新城疫及传染性法氏囊病毒复制的影响[J]. 生物技术通报, 2022, 38(11): 90-96. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||