








生物技术通报 ›› 2023, Vol. 39 ›› Issue (12): 169-178.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0392
任丽1,2( ), 乔舒婷1,2, 葛晨辉1,2, 魏梓桐1,2, 徐晨曦1,2(
), 乔舒婷1,2, 葛晨辉1,2, 魏梓桐1,2, 徐晨曦1,2( )
)
收稿日期:2023-04-23
出版日期:2023-12-26
发布日期:2024-01-11
通讯作者:
徐晨曦,女,博士,副教授,研究方向:菠菜遗传育种及分子生物学;E-mail: chenxixu@shnu.edu.cn作者简介:任丽,女,硕士研究生,研究方向:菠菜遗传育种及分子生物学;E-mail: renlirenli7@163.com
基金资助:
REN Li1,2( ), QIAO Shu-ting1,2, GE Chen-hui1,2, WEI Zi-tong1,2, XU Chen-xi1,2(
), QIAO Shu-ting1,2, GE Chen-hui1,2, WEI Zi-tong1,2, XU Chen-xi1,2( )
)
Received:2023-04-23
Published:2023-12-26
Online:2024-01-11
摘要:
八氢番茄红素合成酶(phytoene synthase, PSY)是植物类胡萝卜素合成途径中的第一个限速酶,对植物的生长发育具有重要作用。为了解菠菜(Spinacia oleracea L.)中PSY基因家族功能,本研究通过生物信息学方法对菠菜PSY基因家族成员的数目、结构、启动子元件、氨基酸特性和基因进化等进行鉴定分析,并利用实时荧光定量PCR技术(RT-qPCR)分析其在菠菜组织中及响应红蓝光比例变化的表达模式,同时测定R1B3(红光∶蓝光=1∶3)和R3B1(红光∶蓝光=3∶1)处理后菠菜总类胡萝卜素含量,进行SoPSYs基因表达量与总类胡萝卜素含量变化的相关性分析。结果显示,菠菜共有4个PSY基因,含有SQS_PSY保守结构域,SoPSY蛋白为亲水性蛋白。菠菜PSY1、PSY2和苋科的甜菜进化关系较近,PSY3、PSY4与番茄PSY3有较近的亲缘关系。菠菜PSY基因启动子序列上存在多种光响应和植物激素相关元件。SoPSY基因具有组织表达特异性,SoPSY1、SoPSY2、SoPSY3仅在叶片中表达,SoPSY4主要在根中表达。菠菜类胡萝卜素含量在R1B3和R3B1处理后均下降。不同基因型菠菜中,各SoPSY基因响应红蓝光比例变化的表达模式具有差异性,其中SoPSY1的表达在栽培类型菠菜中无显著变化,SoPSY2的表达模式在两种类型菠菜中相反,SoPSY3在野生型与栽培型菠菜中表达模式一致,且这3个基因的相对表达水平与总类胡萝卜素含量变化密切相关。本研究为深入了解PSY基因在菠菜中的生物学功能,以及响应红蓝光比例变化的调控机理奠定了基础。
任丽, 乔舒婷, 葛晨辉, 魏梓桐, 徐晨曦. 菠菜PSY基因家族的鉴定与表达分析[J]. 生物技术通报, 2023, 39(12): 169-178.
REN Li, QIAO Shu-ting, GE Chen-hui, WEI Zi-tong, XU Chen-xi. Identification and Expression Analysis of Spinach PSY Gene Family[J]. Biotechnology Bulletin, 2023, 39(12): 169-178.
| 引物名称 Primer name | 上游引物 Upstream primer(5'-3') | 下游引物 Downstream primer(5'-3') |
|---|---|---|
| 18S | CTGAGAAACGGCTACCACA | CCCAAGGTCCAACTACGAG |
| SoPSY1 | CGGTGTAGTGAAGTTTGTGCT | TGCCCAAATGGCTTTTCTCTTT |
| SoPSY2 | CGGTGTAGTGAAGTTTGTGCTGA | TATAGCCCAAATGGCTTTTCGCTTC |
| SoPSY3 | GCTCTTTCCTTGGGTACTGCT | GAGCTCGTCTTGTGGAAGGT |
| SoPSY4 | ATGGCCGAACTTCGTTGATG | CCCTTATTGGATAGAGCAAGAAAGC |
表1 实时荧光定量PCR引物
Table 1 Primers used for quantitative real-time PCR
| 引物名称 Primer name | 上游引物 Upstream primer(5'-3') | 下游引物 Downstream primer(5'-3') |
|---|---|---|
| 18S | CTGAGAAACGGCTACCACA | CCCAAGGTCCAACTACGAG |
| SoPSY1 | CGGTGTAGTGAAGTTTGTGCT | TGCCCAAATGGCTTTTCTCTTT |
| SoPSY2 | CGGTGTAGTGAAGTTTGTGCTGA | TATAGCCCAAATGGCTTTTCGCTTC |
| SoPSY3 | GCTCTTTCCTTGGGTACTGCT | GAGCTCGTCTTGTGGAAGGT |
| SoPSY4 | ATGGCCGAACTTCGTTGATG | CCCTTATTGGATAGAGCAAGAAAGC |
| 基因名称 Gene name | 基因ID Gene ID | 染色体编号 Chrom No. | 氨基酸长 Length/aa | 分子质量 Molecular weight/kD | 等电点 pI | 重力值 GRAVY | 亚细胞定位 Location |
|---|---|---|---|---|---|---|---|
| SoPSY1 | Spo03700 | SpoScf_03339 | 418 | 47.15 | 8.88 | -0.262 | Chloroplast |
| SoPSY2 | Spo13982 | SpoScf_02365 | 418 | 47.14 | 8.89 | -0.262 | Chloroplast |
| SoPSY3 | Spo00660 | chr4 | 359 | 41.48 | 9.17 | -0.494 | Chloroplast |
| SoPSY4 | Spo15086 | chr4 | 351 | 40.24 | 9.11 | -0.363 | Chloroplast |
表2 菠菜PSY基因家族的鉴定及理化性质
Table 2 Identification and characteristic features of PSY gene family in spinach
| 基因名称 Gene name | 基因ID Gene ID | 染色体编号 Chrom No. | 氨基酸长 Length/aa | 分子质量 Molecular weight/kD | 等电点 pI | 重力值 GRAVY | 亚细胞定位 Location |
|---|---|---|---|---|---|---|---|
| SoPSY1 | Spo03700 | SpoScf_03339 | 418 | 47.15 | 8.88 | -0.262 | Chloroplast |
| SoPSY2 | Spo13982 | SpoScf_02365 | 418 | 47.14 | 8.89 | -0.262 | Chloroplast |
| SoPSY3 | Spo00660 | chr4 | 359 | 41.48 | 9.17 | -0.494 | Chloroplast |
| SoPSY4 | Spo15086 | chr4 | 351 | 40.24 | 9.11 | -0.363 | Chloroplast |
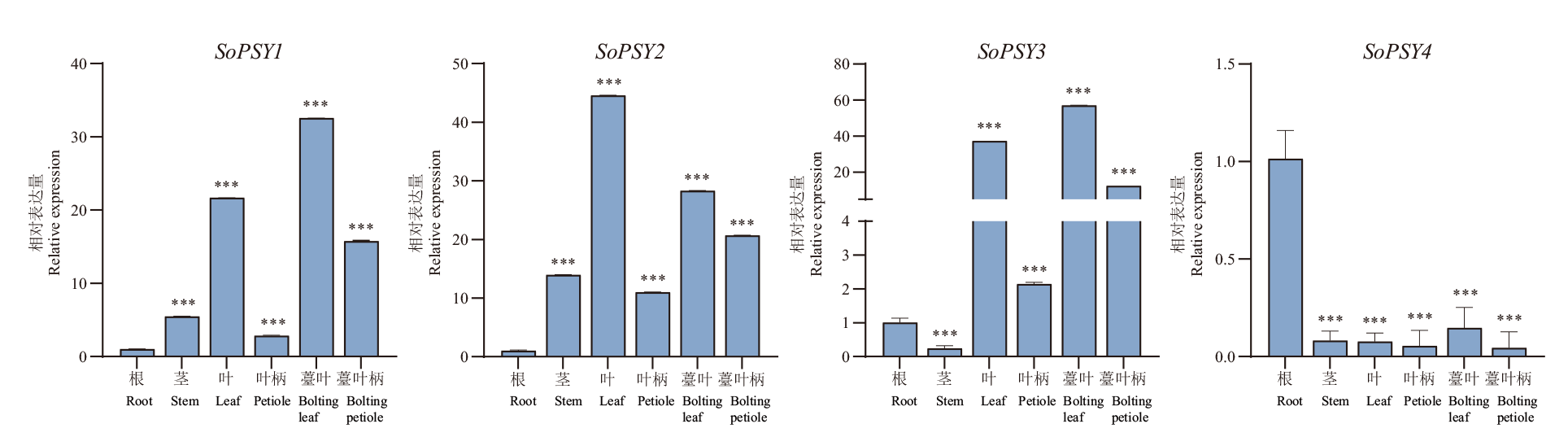
图4 菠菜不同组织中PSY基因的相对表达量 图中误差线表示标准偏差; *P<0.05,** P<0.01和*** P<0.001;下同
Fig. 4 Relative expressions of PSY genes in the different tissues of spinach The error line in the figure refers to the standard deviation ; *P<0.05, ** P<0.01 and *** P<0.001. The same below
| 处理 Treatment | 含量变化量 Content variation | 24 h基因相对表达量24 h relative expression | 12 d基因相对表达量12 d relative expression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SoPSY1 | SoPSY2 | SoPSY3 | SoPSY4 | SoPSY1 | SoPSY2 | SoPSY3 | SoPSY4 | ||||
| CK | US362 | -0.849 | 0.615 | -0.102 | -1.000** | 0.999* | -0.593 | 0.745 | -0.999* | ||
| KS4 | -0.362 | -0.475 | -0.104 | -0.920 | -0.754 | -0.686 | -0.348 | -0.146 | |||
| R1B3 | US362 | -0.058 | 0.749 | 0.673 | 0.570 | -0.042 | 0.999* | 0.953 | 0.765 | ||
| KS4 | -0.476 | -0.490 | -0.479 | -0.726 | -0.978 | -0.955 | -0.011 | 0.011 | |||
| R3B1 | US362 | -0.999* | -0.485 | 0.954 | -0.485 | -0.970 | 0.492 | -0.970 | -0.338 | ||
| KS4 | 0.128 | -0.380 | 0.534 | 0.765 | -0.697 | 0.260 | -0.061 | -0.156 | |||
表3 红蓝光处理下菠菜总类胡萝卜素含量变化与PSY基因相对表达量的相关性分析
Table 3 Correlation analysis of total carotenoid content and PSY gene relative expression in spinach under red and blue light treatment
| 处理 Treatment | 含量变化量 Content variation | 24 h基因相对表达量24 h relative expression | 12 d基因相对表达量12 d relative expression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SoPSY1 | SoPSY2 | SoPSY3 | SoPSY4 | SoPSY1 | SoPSY2 | SoPSY3 | SoPSY4 | ||||
| CK | US362 | -0.849 | 0.615 | -0.102 | -1.000** | 0.999* | -0.593 | 0.745 | -0.999* | ||
| KS4 | -0.362 | -0.475 | -0.104 | -0.920 | -0.754 | -0.686 | -0.348 | -0.146 | |||
| R1B3 | US362 | -0.058 | 0.749 | 0.673 | 0.570 | -0.042 | 0.999* | 0.953 | 0.765 | ||
| KS4 | -0.476 | -0.490 | -0.479 | -0.726 | -0.978 | -0.955 | -0.011 | 0.011 | |||
| R3B1 | US362 | -0.999* | -0.485 | 0.954 | -0.485 | -0.970 | 0.492 | -0.970 | -0.338 | ||
| KS4 | 0.128 | -0.380 | 0.534 | 0.765 | -0.697 | 0.260 | -0.061 | -0.156 | |||
| [1] | Águila Ruiz-Sola M, Rodríguez-Concepción M. Carotenoid biosynthesis in Arabidopsis: a colorful pathway[J]. Arabidopsis Book, 2012, 10: e0158. |
| [2] |
Nisar N, Li L, Lu S, et al. Carotenoid metabolism in plants[J]. Mol Plant, 2015, 8(1): 68-82.
doi: 10.1016/j.molp.2014.12.007 pmid: 25578273 |
| [3] |
Quian-Ulloa R, Stange C. Carotenoid biosynthesis and plastid development in plants: the role of light[J]. Int J Mol Sci, 2021, 22(3): 1184.
doi: 10.3390/ijms22031184 URL |
| [4] |
Sun TH, Yuan H, Cao HB, et al. Carotenoid metabolism in plants: the role of plastids[J]. Mol Plant, 2018, 11(1): 58-74.
doi: S1674-2052(17)30273-3 pmid: 28958604 |
| [5] | Hashimoto H, Uragami C, Cogdell RJ. Carotenoids and photosynthesis[M]// Carotenoids in Nature. Cham: Springer, 2016: 111-139. |
| [6] |
Havaux M. Carotenoid oxidation products as stress signals in plants[J]. Plant J, 2014, 79(4): 597-606.
doi: 10.1111/tpj.2014.79.issue-4 URL |
| [7] |
Johnson MP, Havaux M, Triantaphylidès C, et al. Elevated Zeaxanthin bound to oligomeric LHCII enhances the resistance of Arabidopsis to photooxidative stress by a lipid-protective, antioxidant mechanism[J]. J Biol Chem, 2007, 282(31): 22605-22618.
doi: 10.1074/jbc.M702831200 URL |
| [8] |
Matusova R, Rani K, Verstappen FWA, et al. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway[J]. Plant Physiol, 2005, 139(2): 920-934.
doi: 10.1104/pp.105.061382 URL |
| [9] |
Al-Babili S, Bouwmeester HJ. Strigolactones, a novel carotenoid-derived plant hormone[J]. Annu Rev Plant Biol, 2015, 66: 161-186.
doi: 10.1146/annurev-arplant-043014-114759 pmid: 25621512 |
| [10] |
Eggersdorfer M, Wyss A. Carotenoids in human nutrition and health[J]. Arch Biochem Biophys, 2018, 652: 18-26.
doi: S0003-9861(18)30165-6 pmid: 29885291 |
| [11] | 李静文. 芹菜类胡萝卜素的积累及相关合成基因的功能初步分析[D]. 南京: 南京农业大学, 2019. |
| Li JW. Carotenoids accumulation and preliminary funct analysis of related biosynthsis genes in celery[D]. Nanjing: Nanjing Agricultural University, 2019. | |
| [12] | Zimmer JP, Jr Hammond BR. Possible influences of lutein and Zeaxanthin on the developing retina[J]. Clin Ophthalmol, 2007, 1(1): 25-35. |
| [13] |
Vargas-Murga L, de Rosso VV, Mercadante AZ, et al. Fruits and vegetables in the Brazilian Household Budget Survey(2008-2009): carotenoid content and assessment of individual carotenoid intake[J]. J Food Compos Anal, 2016, 50: 88-96.
doi: 10.1016/j.jfca.2016.05.012 URL |
| [14] |
Dias MG, Borge GIA, Kljak K, et al. European database of carotenoid levels in foods. Factors affecting carotenoid content[J]. Foods, 2021, 10(5): 912.
doi: 10.3390/foods10050912 URL |
| [15] |
Shumskaya M, Wurtzel ET. The carotenoid biosynthetic pathway: thinking in all dimensions[J]. Plant Sci, 2013, 208: 58-63.
doi: 10.1016/j.plantsci.2013.03.012 pmid: 23683930 |
| [16] |
Yuan H, Zhang JX, Nageswaran D, et al. Carotenoid metabolism and regulation in horticultural crops[J]. Hortic Res, 2015, 2: 15036.
doi: 10.1038/hortres.2015.36 |
| [17] |
Cazzonelli CI, Pogson BJ. Source to sink: regulation of carotenoid biosynthesis in plants[J]. Trends Plant Sci, 2010, 15(5): 266-274.
doi: 10.1016/j.tplants.2010.02.003 pmid: 20303820 |
| [18] |
Zhou XS, Rao S, Wrightstone E, et al. Phytoene synthase: the key rate-limiting enzyme of carotenoid biosynthesis in plants[J]. Front Plant Sci, 2022, 13: 884720.
doi: 10.3389/fpls.2022.884720 URL |
| [19] |
Sun TH, Tadmor Y, Li L. Pathways for carotenoid biosynthesis, degradation, and storage[J]. Methods Mol Biol, 2020, 2083: 3-23.
doi: 10.1007/978-1-4939-9952-1_1 pmid: 31745909 |
| [20] |
Hirschberg J. Carotenoid biosynthesis in flowering plants[J]. Curr Opin Plant Biol, 2001, 4(3): 210-218.
doi: 10.1016/s1369-5266(00)00163-1 pmid: 11312131 |
| [21] |
Tran D, Haven J, Qiu WG, et al. An update on carotenoid biosynthesis in algae: phylogenetic evidence for the existence of two classes of phytoene synthase[J]. Planta, 2009, 229(3): 723-729.
doi: 10.1007/s00425-008-0866-2 pmid: 19066941 |
| [22] |
Scolnik PA, Bartley GE. Nucleotide sequence of an Arabidopsis cDNA for phytoene synthase[J]. Plant Physiol, 1994, 104(4): 1471-1472.
pmid: 8016277 |
| [23] |
Welsch R, Wüst F, Bär C, et al. A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes[J]. Plant Physiol, 2008, 147(1): 367-380.
doi: 10.1104/pp.108.117028 pmid: 18326788 |
| [24] |
Li FQ, Vallabhaneni R, Yu J, et al. The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance[J]. Plant Physiol, 2008, 147(3): 1334-1346.
doi: 10.1104/pp.108.122119 pmid: 18508954 |
| [25] |
Stauder R, Welsch R, Camagna M, et al. Strigolactone levels in dicot roots are determined by an ancestral symbiosis-regulated clade of the PHYTOENE SYNTHASE gene family[J]. Front Plant Sci, 2018, 9: 255.
doi: 10.3389/fpls.2018.00255 pmid: 29545815 |
| [26] |
Dibari B, Murat F, Chosson A, et al. Deciphering the genomic structure, function and evolution of carotenogenesis related phytoene synthases in grasses[J]. BMC Genomics, 2012, 13: 221.
doi: 10.1186/1471-2164-13-221 pmid: 22672222 |
| [27] |
Ampomah-Dwamena C, Driedonks N, Lewis D, et al. The Phytoene synthase gene family of apple(Malus×domestica)and its role in controlling fruit carotenoid content[J]. BMC Plant Biol, 2015, 15: 185.
doi: 10.1186/s12870-015-0573-7 pmid: 26215656 |
| [28] |
López-Emparán A, Quezada-Martinez D, Zúñiga-Bustos M, et al. Functional analysis of the Brassica napus L. phytoene synthase(PSY)gene family[J]. PLoS One, 2014, 9(12): e114878.
doi: 10.1371/journal.pone.0114878 URL |
| [29] | 孟纯阳. 辣椒PSY基因家族分析及CaPSY1功能研究[D]. 郑州: 郑州大学, 2019. |
| Meng CY. Genome-wide identification of PSY gene family and function analysis of CaPSY1 in pepper(Capsicum annuum L.)[D]. Zhengzhou: Zhengzhou University, 2019. | |
| [30] |
Cao HB, Luo HM, Yuan H, et al. A neighboring aromatic-aromatic amino acid combination governs activity divergence between tomato phytoene synthases[J]. Plant Physiol, 2019, 180(4): 1988-2003.
doi: 10.1104/pp.19.00384 pmid: 31221734 |
| [31] |
Frede K, Schreiner M, Zrenner R, et al. Carotenoid biosynthesis of pak choi(Brassica rapa ssp. chinensis)sprouts grown under different light-emitting diodes during the diurnal course[J]. Photochem Photobiol Sci, 2018, 17(10): 1289-1300.
doi: 10.1039/c8pp00136g URL |
| [32] |
Bou-Torrent J, Toledo-Ortiz G, Ortiz-Alcaide M, et al. Correction to: regulation of carotenoid biosynthesis by shade relies on specific subsets of antagonistic transcription factors and cofactors[J]. Plant Physiol, 2022, 189(2): 1171.
doi: 10.1093/plphys/kiac120 URL |
| [1] | 吴巧茵, 施友志, 李林林, 彭政, 谭再钰, 刘利平, 张娟, 潘勇. 类胡萝卜素降解菌株的原位筛选及其在雪茄提质增香中的应用[J]. 生物技术通报, 2023, 39(9): 192-201. |
| [2] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [3] | 叶云芳, 田清尹, 施婷婷, 王亮, 岳远征, 杨秀莲, 王良桂. 植物中β-紫罗兰酮生物合成及调控研究进展[J]. 生物技术通报, 2023, 39(8): 91-105. |
| [4] | 张和臣, 袁欣, 高杰, 王校晨, 王慧娟, 李艳敏, 王利民, 符真珠, 李保印. 植物花瓣呈色机理及花色分子育种[J]. 生物技术通报, 2023, 39(5): 23-31. |
| [5] | 张志霞, 李天培, 曾虹, 朱稀贤, 杨天雄, 马斯楠, 黄磊. 冰冷杆菌PG-2的基因组测序及生物信息学分析[J]. 生物技术通报, 2023, 39(3): 290-300. |
| [6] | 罗皓天, 王龙, 王禹茜, 王月, 李佳祯, 杨梦珂, 张杰, 邓欣, 王红艳. 青狗尾草RNAi途径相关基因的全基因组鉴定和表达分析[J]. 生物技术通报, 2023, 39(1): 175-186. |
| [7] | 周琳, 梁轩铭, 赵磊. 天然类胡萝卜素的生物合成研究进展[J]. 生物技术通报, 2022, 38(7): 119-127. |
| [8] | 王慧, 马艺文, 乔正浩, 常彦彩, 术琨, 丁海萍, 聂永心, 潘光堂. AOX基因家族的结构和功能特征分析[J]. 生物技术通报, 2022, 38(7): 160-170. |
| [9] | 田清尹, 岳远征, 申慧敏, 潘多, 杨秀莲, 王良桂. 植物观赏器官中类胡萝卜素代谢调控的研究进展[J]. 生物技术通报, 2022, 38(12): 35-46. |
| [10] | 曹映辉, 胡美娟, 童妍, 张燕萍, 赵凯, 彭东辉, 周育真. 建兰ABC基因家族鉴定及其在花发育过程中的表达模式分析[J]. 生物技术通报, 2022, 38(11): 162-174. |
| [11] | 孔谦, 黄文洁, 吴绍文, 李坤, 张名位, 晏石娟. 一种同时测定十种类胡萝卜素的液相色谱方法的建立[J]. 生物技术通报, 2022, 38(11): 80-89. |
| [12] | 高玲, 王斐, 谢双全, 陈喜凤, 沈海涛, 李鸿彬. 乌拉尔甘草CBL基因家族的鉴定与表达分析[J]. 生物技术通报, 2021, 37(4): 18-27. |
| [13] | 王悦, 欧阳丹, 汤伟, 刘仕博, 顾燕, 何增国. 六株红酵母抗氧化活性的研究[J]. 生物技术通报, 2020, 36(10): 156-164. |
| [14] | 李爱国, 李积铭, 李和平, 刘桂华, 宋聪敏, 武军艳. 适宜河北省旱寒区的冬油菜品种的筛选[J]. 生物技术通报, 2020, 36(1): 95-100. |
| [15] | 王雪寒, 马强, 田媛, 胡靖, 刘惠荣. 内蒙古呼伦贝尔地区的可培养黏细菌及其抗菌活性[J]. 生物技术通报, 2019, 35(9): 224-233. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||