








生物技术通报 ›› 2024, Vol. 40 ›› Issue (2): 160-171.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0856
龚丽丽1,2( ), 余花2, 杨杰2, 陈天池2, 赵双滢2, 吴月燕1,2(
), 余花2, 杨杰2, 陈天池2, 赵双滢2, 吴月燕1,2( )
)
收稿日期:2023-09-04
出版日期:2024-02-26
发布日期:2024-03-13
通讯作者:
吴月燕,女,硕士,教授,研究方向:植物生理与分子生物学;E-mail: wyy2000@zwu.edu.cn作者简介:龚丽丽,女,硕士研究生,研究方向:植物生理与分子生物学;E-mail: gongllstudy@163.com
基金资助:
GONG Li-li1,2( ), YU Hua2, YANG Jie2, CHEN Tian-chi2, ZHAO Shuang-ying2, WU Yue-yan1,2(
), YU Hua2, YANG Jie2, CHEN Tian-chi2, ZHAO Shuang-ying2, WU Yue-yan1,2( )
)
Received:2023-09-04
Published:2024-02-26
Online:2024-03-13
摘要:
【目的】CYP707A是细胞色素P450家族的亚家族成员,所编码的ABA8'-羟化酶是ABA分解代谢的关键酶,对植物生长发育起着重要作用。鉴定葡萄CYP707A基因家族,可为进一步研究葡萄果实成熟机理和生物育种技术提供理论依据。【方法】利用生物学信息方法分析葡萄中CYP707A家族基因的基因结构、蛋白基序、染色体定位、共线性、系统进化关系及启动子区顺式作用元件等,并利用RT-qPCR分析VvCYP707A基因在葡萄不同品种(早熟和中熟)的时空表达模式、组织表达模式以及对外源ABA处理的响应模式,并利用果实瞬时表达技术对Vitvi07g01751进行功能验证。【结果】葡萄基因组鉴定出5个VvCYP707As基因,分布于5条染色体,其中Vitvi02g01269、Vitvi07g01751和Vitvi18g00792为片段复制基因。CYP707A基因家族高度保守,其中,基因结构和蛋白基序相似,亚细胞定位均位于内质网;VvCYP707A启动子区域富含与生长发育和激素响应相关的顺式作用元件,均含有ABA激素响应元件;通过RT-qPCR检测基因的表达模式发现,VvCYP707A基因具有时空特异性和组织特异性,主要在果实的幼果期与膨大期和茎、叶组织中高表达,其中Vitvi07g01751基因在早熟品种‘甬早红’膨大期果实中特异性高表达,且在两个品种间的表达模式一致,随着果实的成熟其表达先升高后降低。外源ABA处理下VvCYP707A分为两种表达模式,在浆果肉中外源ABA可诱导5个VvCYP707A基因高表达,在浆果皮中会显著抑制VvCYP707A基因的表达;在葡萄果实中瞬时表达Vitvi07g01751,葡萄的糖度积累缓慢,显著增加编码ABA的受体VvPYL4基因表达量,推测了Vitvi07g01751介导ABA调控葡萄果实成熟的机制。【结论】研究结果揭示了葡萄Vitvi07g01751在果实成熟中起着重要作用。
龚丽丽, 余花, 杨杰, 陈天池, 赵双滢, 吴月燕. 葡萄CYP707A基因家族的鉴定及对果实成熟的功能验证[J]. 生物技术通报, 2024, 40(2): 160-171.
GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening[J]. Biotechnology Bulletin, 2024, 40(2): 160-171.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| Vitvi07g01751-F | ATGCTGCTGAAGAAGCCACA |
| Vitvi07g01751-R | TCAGCATTCCTTCCAAAACTTAGC |
| 35S:VvCYP707A-F | ACGAGCTCGGTACCCGGGATGCTGCTGAAGAAGCCACAG |
| 35S:VvCYP707A-R | GGGCGAATTGGTCGACGCATTCCTTCCAAAACTTAGCAGG |
表1 引物信息
Table 1 Primer information
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| Vitvi07g01751-F | ATGCTGCTGAAGAAGCCACA |
| Vitvi07g01751-R | TCAGCATTCCTTCCAAAACTTAGC |
| 35S:VvCYP707A-F | ACGAGCTCGGTACCCGGGATGCTGCTGAAGAAGCCACAG |
| 35S:VvCYP707A-R | GGGCGAATTGGTCGACGCATTCCTTCCAAAACTTAGCAGG |
| 基因Gene | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| Vitvi02g01269 | F: GTGTTCTGCAAGCTGTCACG | R: TGGCATCTTCCCAAGTCAGAG |
| Vitvi03g00508 | F: ATCGGCCTTTGACATGGGAT | R: AGTGGCATCACCTTCCAACC |
| Vitvi06g00498 | F: GCAGCTTCTCAACGAGACTTTCAGG | R: GAATTCCCAGCAATCCACCTCCTTG |
| Vitvi07g01751 | F: CTCTGCGGCTCTATTCCCAG | R: CTTGGTGGGTATGTGGGCTT |
| Vitvi18g00792 | F: TTGTCCTGGAAATGAGCTTGCCAAG | R: CAATTCCACCTTGTGATCCCACCAC |
| Actin | F: CAAGAGAAACCATCCCTAGCTG | R: TCAATCTGTCTAGGAAAGGAAG |
| VvPYL1 | F: CAAGAGAAACCATCCCTAGCTG | R: TCAATCTGTCTAGGAAAGGAAG |
| VvPYL2 | F: TCCCTACTGTCTGGTCCGTC | R: CCAATCTCTCCGTGCTGGTC |
| VvPYL3 | F: GACGAGTACATCCGCAGACA | R: TGATGTGCTTGACGAGAGAGG |
| VvPYL4 | F: GTGCTCTTCCCTACTCGCTC | R: GAAGCGATGAACGACGGAAC |
| VvPYL5 | F: ATAGACGGCTGTGATGGGTG | R: TGAACTGCACTGGTTCTCCC |
| VvPYL6 | F: CCACTACCAGCACTGAAAGGT | R: TCGTCCTTGGTGTTTCCCTC |
表2 RT-qPCR所用引物
Table 2 Primers for RT-qPCR
| 基因Gene | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') |
|---|---|---|
| Vitvi02g01269 | F: GTGTTCTGCAAGCTGTCACG | R: TGGCATCTTCCCAAGTCAGAG |
| Vitvi03g00508 | F: ATCGGCCTTTGACATGGGAT | R: AGTGGCATCACCTTCCAACC |
| Vitvi06g00498 | F: GCAGCTTCTCAACGAGACTTTCAGG | R: GAATTCCCAGCAATCCACCTCCTTG |
| Vitvi07g01751 | F: CTCTGCGGCTCTATTCCCAG | R: CTTGGTGGGTATGTGGGCTT |
| Vitvi18g00792 | F: TTGTCCTGGAAATGAGCTTGCCAAG | R: CAATTCCACCTTGTGATCCCACCAC |
| Actin | F: CAAGAGAAACCATCCCTAGCTG | R: TCAATCTGTCTAGGAAAGGAAG |
| VvPYL1 | F: CAAGAGAAACCATCCCTAGCTG | R: TCAATCTGTCTAGGAAAGGAAG |
| VvPYL2 | F: TCCCTACTGTCTGGTCCGTC | R: CCAATCTCTCCGTGCTGGTC |
| VvPYL3 | F: GACGAGTACATCCGCAGACA | R: TGATGTGCTTGACGAGAGAGG |
| VvPYL4 | F: GTGCTCTTCCCTACTCGCTC | R: GAAGCGATGAACGACGGAAC |
| VvPYL5 | F: ATAGACGGCTGTGATGGGTG | R: TGAACTGCACTGGTTCTCCC |
| VvPYL6 | F: CCACTACCAGCACTGAAAGGT | R: TCGTCCTTGGTGTTTCCCTC |

图1 葡萄CYP707A基因家族氨基酸比对(A)、共线性分析(B)与染色体定位(C)
Fig. 1 Amino acid alignment(A), collinearity analysis(B)and chromosome localization(C)of grape CYP707A gene family
| 基因ID Gene ID | 氨基酸长度 Amino acid length/aa | 分子量大小 Molecular weight/kD | 等电点 pI | 亚细胞定位 Subcellular location |
|---|---|---|---|---|
| Vitvi02g01269 | 470 | 53 118.2 | 9.03 | 内质网 |
| Vitvi03g00508 | 479 | 54 538.9 | 9.68 | 内质网 |
| Vitvi06g00498 | 488 | 55 496.0 | 10.00 | 内质网 |
| Vitvi07g01751 | 447 | 50 689.3 | 9.50 | 内质网 |
| Vitvi18g00792 | 470 | 53 448.7 | 10.08 | 内质网 |
表3 葡萄CYP707A蛋白相关信息
Table 3 Information of CYP707A proteins in grape
| 基因ID Gene ID | 氨基酸长度 Amino acid length/aa | 分子量大小 Molecular weight/kD | 等电点 pI | 亚细胞定位 Subcellular location |
|---|---|---|---|---|
| Vitvi02g01269 | 470 | 53 118.2 | 9.03 | 内质网 |
| Vitvi03g00508 | 479 | 54 538.9 | 9.68 | 内质网 |
| Vitvi06g00498 | 488 | 55 496.0 | 10.00 | 内质网 |
| Vitvi07g01751 | 447 | 50 689.3 | 9.50 | 内质网 |
| Vitvi18g00792 | 470 | 53 448.7 | 10.08 | 内质网 |

图2 CYP707A蛋白的系统进化树 MEGA X 软件构建24个CYP707A氨基酸序列系统进化树,其中包括5个葡萄、2个番茄、4个拟南芥、2个胡萝卜、4个水稻、2个苹果、2个马铃薯和4个柑橘CYP707A蛋白。葡萄的VvCYP707A蛋白用红色和红色星形标记
Fig. 2 Phylogenetic tree analysis of CYP707A protein The phylogenetic tree was constructed with the full-length amino acid sequences of the 24 CYP707A using MEGA X. The analysis included 5 Vitis vinifera VvCYP707A proteins, 2 Solanum lycopersicum SlCYP707A proteins, 4 Arabidopsis thaliana AtCYP707A proteins, 2 Daucus carota DcCYP707A proteins, 4 Oryza sativa OsCYP707A proteins, 2 Malus domestica MdCYP707A proteins, 2 Solanum tuberosum StCYP707A proteins and 4 Citrus CsCYP707A proteins. The VvCYP707A proteins in V. vinifera were marked with red color and red stars
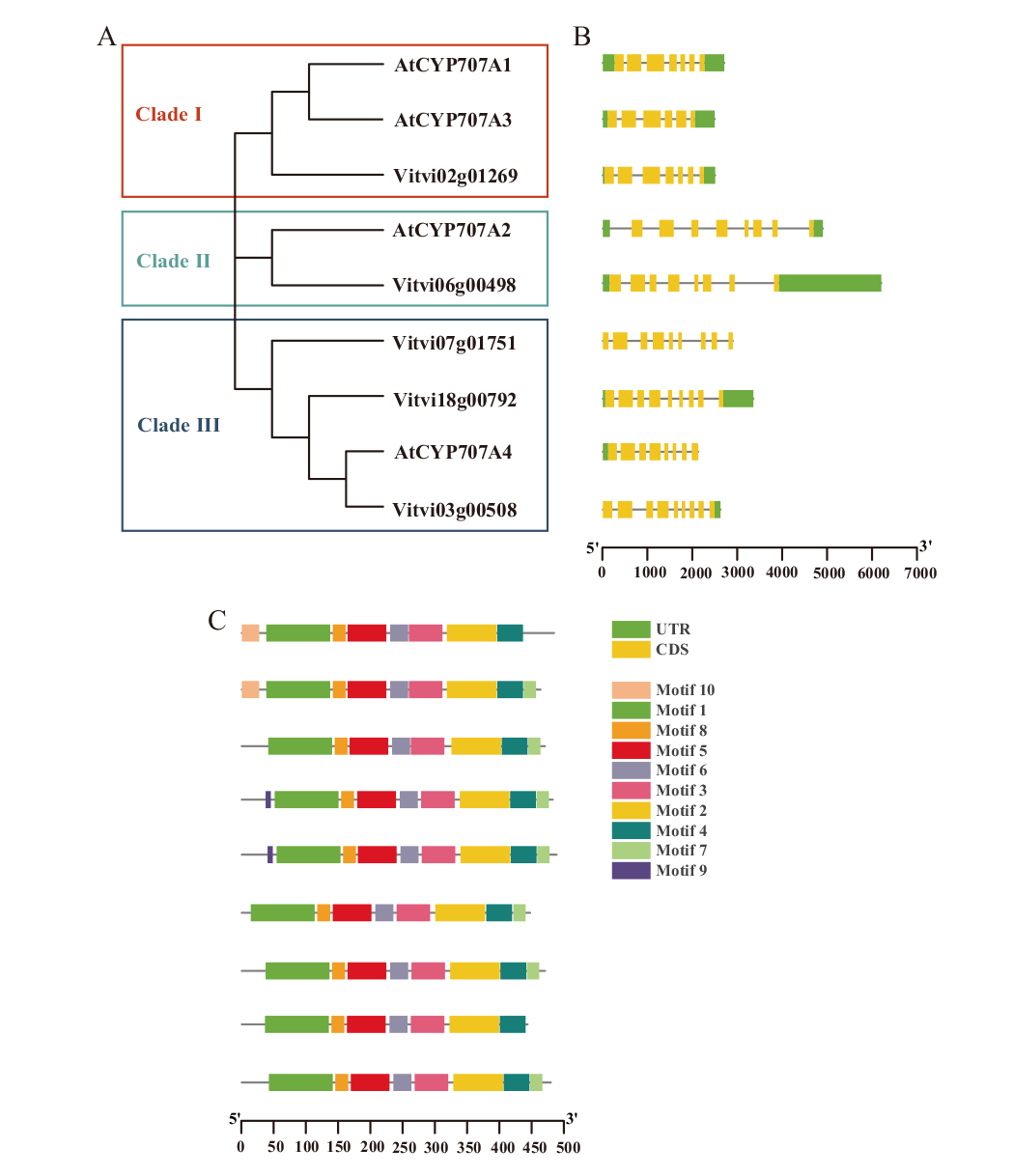
图3 拟南芥和葡萄CYP707As的系统发育关系(A)、基因结构(B)及保守基序分析(C)
Fig. 3 Phylogenetic relationships (A), gene structure (B) and conserved motif analyses (C) of CYP707As from Arabidopsis and V. vinifera

图5 两种葡萄果实发育的生理参数及VvCYP707As基因相对表达量 A:‘鄞红’和‘甬早红’不同发育时期,S1、S2幼果期,S3膨大期,S4转色期,S5成熟期,下同;B-D:‘鄞红’和‘甬早红’果实单重、果实硬度与可溶性糖含量,所用数据为3个生物学重复的平均值±SD,柱上*表示两个品种间差异显著,*:P<0.05,**:P<0.01,***:P<0.001;E:VvCYP707A在‘鄞红’和‘甬早红’中基因表达变化;不同小写字母代表显著差异(P<0.05);n=3,下同
Fig. 5 Physiological parameters and VvCYP707As related expression related to fruit development in two kinds of V. vinifera A: ‘Yin Hong’ and ‘Yong Zaohong’ at different developmental periods; S1-S2: young fruit stage; S3: expansion stage; S4: veraison stage; S5: mature stage. The same below. B-D: ‘Yin Hong’ and ‘Yong Zaohong’ fruit single weight, fruit hardness and soluble sugar content. The data used are the mean ±SD of the three biological replicates. * on the column indicates significant differences between the two varieties, *: P<0.05, **: P<0.01, ***: P<0.001. E: VvCYP707A gene expression changes in ‘Yin Hong’ and ‘Yong Zaohong’. Different lowercase letters represent a significant difference(P<0.05); n=3, the same below
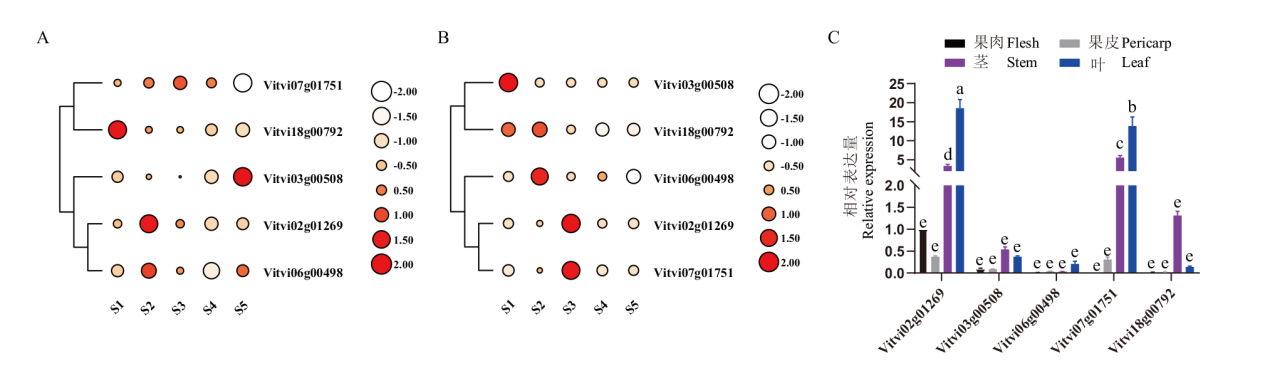
图6 VvCYP707As在‘鄞红’不同组织不同时期表达模式 A:果肉;B:果皮;C:VvCYP707As转色期表达量组织特异性
Fig. 6 VvCYP707As expression patterns in different tissues and different periods in ‘Yin Hong’ A: Flesh; B: pericarp; C: tissue specificity of VvCYP707As expression in veraison stage
| [1] |
Chatterjee A, Dhal S, Pal H. Insight into the regulatory network of miRNA to unravel the ripening physiology of climacteric and non-climacteric fruits[J]. Plant Gene, 2021, 28: 100329.
doi: 10.1016/j.plgene.2021.100329 URL |
| [2] |
Li DM, Pang YJ, Li H, et al. Comparative analysis of the gene expression profile under two cultivation methods reveals the critical role of ABA in grape quality promotion[J]. Sci Hortic, 2021, 281: 109924.
doi: 10.1016/j.scienta.2021.109924 URL |
| [3] |
Kondo S, Sugaya S, Sugawa S, et al. Dehydration tolerance in apple seedlings is affected by an inhibitor of ABA 8'-hydroxylase CYP707A[J]. J Plant Physiol, 2012, 169(3): 234-241.
doi: 10.1016/j.jplph.2011.09.007 URL |
| [4] |
Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism[J]. Annu Rev Plant Biol, 2005, 56: 165-185.
pmid: 15862093 |
| [5] |
Cutler AJ, Krochko JE. Formation and breakdown of ABA[J]. Trends Plant Sci, 1999, 4(12): 472-478.
doi: 10.1016/s1360-1385(99)01497-1 pmid: 10562731 |
| [6] |
Rodriguez PL. Abscisic acid catabolism generates phaseic acid, a molecule able to activate a subset of ABA receptors[J]. Mol Plant, 2016, 9(11): 1448-1450.
doi: S1674-2052(16)30217-9 pmid: 27693497 |
| [7] |
Weng JK, Ye ML, Li B, et al. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity[J]. Cell, 2016, 166(4): 881-893.
doi: 10.1016/j.cell.2016.06.027 URL |
| [8] |
Okamoto M, Kushiro T, Jikumaru Y, et al. ABA 9'-hydroxylation is catalyzed by CYP707A in Arabidopsis[J]. Phytochemistry, 2011, 72(8): 717-722.
doi: 10.1016/j.phytochem.2011.02.004 URL |
| [9] |
Brun G, Thoiron S, Braem L, et al. CYP707As are effectors of karrikin and strigolactone signalling pathways in Arabidopsis thaliana and parasitic plants[J]. Plant Cell Environ, 2019, 42(9): 2612-2626.
doi: 10.1111/pce.v42.9 URL |
| [10] |
Kushiro T, Okamoto M, Nakabayashi K, et al. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8-hydroxylases: key enzymes in ABA catabolism[J]. EMBO J, 2004, 23(7): 1647-1656.
doi: 10.1038/sj.emboj.7600121 pmid: 15044947 |
| [11] |
Sapkota S, Liu JY, Islam MT, et al. Contrasting bloom dates in two apple cultivars linked to differential levels of phytohormones and heat requirements during ecodormancy[J]. Sci Hortic, 2021, 288: 110413.
doi: 10.1016/j.scienta.2021.110413 URL |
| [12] |
Gonzalez HCB, Galli V. The CYP707A gene family in strawberry(Fragaria × ananassa)[J]. Braz Arch Biol Technol, 2021, 64: e21200133.
doi: 10.1590/1678-4324-2021200133 URL |
| [13] |
Ren J, Sun L, Wu JF, et al. Cloning and expression analysis of cDNAs for ABA 8'-hydroxylase during sweet cherry fruit maturation and under stress conditions[J]. J Plant Physiol, 2010, 167(17): 1486-1493.
doi: 10.1016/j.jplph.2010.05.027 URL |
| [14] |
Parwez R, Aftab T, Gill SS, et al. Abscisic acid signaling and crosstalk with phytohormones in regulation of environmental stress responses[J]. Environ Exp Bot, 2022, 199: 104885.
doi: 10.1016/j.envexpbot.2022.104885 URL |
| [15] | 吴月燕, 陈天池, 王立如, 等. 鲜食葡萄新品种‘甬早红’[J]. 园艺学报, 2022, 49(S2): 41-42. |
|
Wu YY, Chen TC, Wang LR, et al. A new table grape cultivar‘Yongzaohong’[J]. Acta Hortic Sin, 2022, 49(S2): 41-42.
doi: 10.16420/j.issn.0513-353x.2022-0550 |
|
| [16] | 张友杰. 以蒽酮分光光度法测定果蔬中的葡萄糖、果糖、蔗糖和淀粉[J]. 分析化学, 1977, 5(3): 167-171. |
| Zhang YJ. Determination of glucose, fructose, sucrose and starch in fruits and vegetables by anthrone spectrophotometry[J]. Chin J Anal Chem, 1977, 5(3): 167-171. | |
| [17] |
Ren C, Zhang Z, Wang Y, et al. Genome-wide identification and characterization of the NF-Y gene family in grape(Vitis vinifera L.)[J]. BMC Genomics, 2016, 17(1): 605.
doi: 10.1186/s12864-016-2989-3 URL |
| [18] |
李敬蕊, 王育博, 解紫薇, 等. 甜瓜PIN基因家族的鉴定及高温胁迫表达分析[J]. 生物技术通报, 2023, 39(5): 192-204.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1196 |
| Li JR, Wang YB, Xie ZW, et al. Identification and expression analysis of PIN gene family in melon under high temperature stress[J]. Biotechnol Bull, 2023, 39(5): 192-204. | |
| [19] |
Rehal PK, Tuan PA, Nguyen TN, et al. Genetic variation of seed dormancy in wheat(Triticum aestivum L.)is mediated by transcriptional regulation of abscisic acid metabolism and signaling[J]. Plant Sci, 2022, 324: 111432.
doi: 10.1016/j.plantsci.2022.111432 URL |
| [20] |
Gupta A, Upadhyay RK, Prabhakar R, et al. SlDREB3, a negative regulator of ABA responses, controls seed germination, fruit size and the onset of ripening in tomato[J]. Plant Sci, 2022, 319: 111249.
doi: 10.1016/j.plantsci.2022.111249 URL |
| [21] |
高真真, 徐功勋, 王东岭, 等. 桃ABA 8'-羟化酶基因PpeCYP707As在拟南芥中过表达的功能分析[J]. 园艺学报, 2018, 45(2): 239-249.
doi: 10.16420/j.issn.0513-353x.2017-0350 |
| Gao ZZ, Xu GX, Wang DL, et al. Functional analysis of peach PpeCYP707As gene in Arabidopsis thaliana overexpressing plants[J]. Acta Hortic Sin, 2018, 45(2): 239-249. | |
| [22] |
Sun WJ, Ma ZT, Liu MY. Cytochrome P450 family: Genome-wide identification provides insights into the rutin synthesis pathway in Tartary buckwheat and the improvement of agricultural product quality[J]. Int J Biol Macromol, 2020, 164: 4032-4045.
doi: 10.1016/j.ijbiomac.2020.09.008 pmid: 32896558 |
| [23] |
Wang H, Umer MJ, Liu F, et al. Genome-wide identification and characterization of CPR5 genes in Gossypium reveals their potential role in trichome development[J]. Front Genet, 2022, 13: 921096.
doi: 10.3389/fgene.2022.921096 URL |
| [24] |
Zhu PP, Cai YX, Yu J, et al. Characterization and expression of abscisic acid signal transduction genes during mulberry fruit ripening[J]. Acta Physiol Plant, 2017, 39(7): 149.
doi: 10.1007/s11738-017-2442-5 URL |
| [25] |
Saito T, Thunyamada S, Wang SS, et al. Exogenous ABA and endogenous ABA affects ‘Kyoho’ grape berry coloration in different pathway[J]. Plant Gene, 2018, 14: 74-82.
doi: 10.1016/j.plgene.2018.05.001 URL |
| [26] |
董昳伶, 肖旭腾, 张敏, 等. 环割对葡萄VvNCED基因的表达和果实成熟的影响[J]. 核农学报, 2022, 36(7): 1339-1349.
doi: 10.11869/j.issn.100-8551.2022.07.1339 |
| Dong YL, Xiao XT, Zhang M, et al. Effect of girdling on the expression of VvNCEDs and fruit ripening in grapes[J]. J Nucl Agric Sci, 2022, 36(7): 1339-1349. | |
| [27] | 邓昌哲, 秦于玲, 李开绵, 等. 外源ABA对木薯叶片β-胡萝卜素合成通路相关基因表达的影响[J]. 热带作物学报, 2017, 38(4): 667-672. |
| Deng CZ, Qin YL, Li KM, et al. Effects of exogenous ABA on the expression of genes associated with β-carotene synthesis pathway in cassava leaves[J]. Chin J Trop Crops, 2017, 38(4): 667-672. | |
| [28] | 王东岭, 杜培勇, 郇蕾, 等. 桃CYP707A家族基因的克隆以及表达分析[J]. 植物生理学报, 2016, 52(5): 659-668. |
| Wang DL, Du PY, Huan L, et al. Molecular cloning and expression analysis of CYP707A gene family in peach[J]. Plant Physiol J, 2016, 52(5): 659-668. | |
| [29] |
Saito S, Hirai N, Matsumoto C, et al. Arabidopsis CYP707As encode(+)-abscisic acid 8'-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid[J]. Plant Physiol, 2004, 134(4): 1439-1449.
doi: 10.1104/pp.103.037614 URL |
| [30] |
Mou WS, Li DD, Bu JW, et al. Comprehensive analysis of ABA effects on ethylene biosynthesis and signaling during tomato fruit ripening[J]. PLoS One, 2016, 11(4): e0154072.
doi: 10.1371/journal.pone.0154072 URL |
| [31] | 刘雪梅. 新疆玛纳斯河流域酿酒葡萄成熟度指标与葡萄酒质量关系的研究[D]. 杨凌: 西北农林科技大学, 2008. |
| Liu XM. The research on wine qulity and grape maturity in the basin of the manas river, Xinjiang[D]. Yangling: Northwest A & F University, 2008. |
| [1] | 路喻丹, 刘晓驰, 冯新, 陈桂信, 陈义挺. 猕猴桃BBX基因家族成员鉴定与转录特征分析[J]. 生物技术通报, 2024, 40(2): 172-182. |
| [2] | 赵光绪, 杨合同, 邵晓波, 崔志豪, 刘红光, 张杰. 一株高效溶磷产红青霉培养条件优化及其溶磷特性[J]. 生物技术通报, 2023, 39(9): 71-83. |
| [3] | 宋志忠, 徐维华, 肖慧琳, 唐美玲, 陈景辉, 管雪强, 刘万好. 酿酒葡萄铁调节转运蛋白基因VvIRT1的克隆、表达与功能[J]. 生物技术通报, 2023, 39(8): 234-240. |
| [4] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [5] | 董聪, 高庆华, 王玥, 罗同阳, 王庆庆. 基于联合策略提高FAD依赖的葡萄糖脱氢酶的酵母表达[J]. 生物技术通报, 2023, 39(6): 316-324. |
| [6] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [7] | 李敬蕊, 王育博, 解紫薇, 李畅, 吴晓蕾, 宫彬彬, 高洪波. 甜瓜PIN基因家族的鉴定及高温胁迫表达分析[J]. 生物技术通报, 2023, 39(5): 192-204. |
| [8] | 陈晓萌, 张雪静, 张欢, 张宝江, 苏艳. 重组牛乳源金黄色葡萄球菌GapC蛋白优势B细胞抗原表位的预测和筛选[J]. 生物技术通报, 2023, 39(5): 306-313. |
| [9] | 赵赛赛, 张小丹, 贾晓妍, 陶大炜, 刘可玉, 宁喜斌. 高产硝酸盐还原酶Staphylococcus simulans ZSJ6的复合诱变选育及其酶学性质研究[J]. 生物技术通报, 2023, 39(4): 103-113. |
| [10] | 赖瑞联, 冯新, 高敏霞, 路喻丹, 刘晓驰, 吴如健, 陈义挺. 猕猴桃过氧化氢酶基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2023, 39(4): 136-147. |
| [11] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| [12] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [13] | 杨岚, 张晨曦, 樊学伟, 王阳光, 王春秀, 李文婷. 鸡 BMP15 基因克隆、表达模式及启动子活性分析[J]. 生物技术通报, 2023, 39(4): 304-312. |
| [14] | 宋海娜, 吴心桐, 杨鲁豫, 耿喜宁, 张华敏, 宋小龙. 葱鳞葡萄孢菌诱导下韭菜RT-qPCR内参基因的筛选和验证[J]. 生物技术通报, 2023, 39(3): 101-115. |
| [15] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||