








生物技术通报 ›› 2024, Vol. 40 ›› Issue (4): 159-166.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0962
收稿日期:2023-10-11
出版日期:2024-04-26
发布日期:2024-04-30
通讯作者:
李丽霞,女,博士,讲师,研究方向:园艺植物生物技术与遗传改良;E-mail: lilixia222@126.com作者简介:肖雅茹,女,硕士研究生,研究方向:园艺植物生物技术与遗传改良;E-mail: cucumberteam2021@126.com
基金资助:
XIAO Ya-ru( ), JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia(
), JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia( )
)
Received:2023-10-11
Published:2024-04-26
Online:2024-04-30
摘要:
【目的】ERF转录因子是AP2/ERF基因家族的一个亚族,在植物生长发育和抵抗外界胁迫方面具有重要作用,克隆CsERF025-like(CsERF025L)并分析其表达模式,为黄瓜CsERF025L功能研究奠定基础。【方法】通过RT-PCR从黄瓜茎尖中克隆CsERF025L,对CsERF025L进行蛋白理化性质、亚细胞定位、互作蛋白等生物信息学分析,采用实时荧光定量PCR分析CsERF025L在黄瓜中的表达模式。【结果】CsERF025L编码序列为510 bp,编码169个氨基酸,具有一个AP2/ERF结构域,属于AP2/ERF家族的ERF亚家族。CsERF025L蛋白是一种不稳定的疏水性蛋白,位于细胞核,蛋白CsERF025L与P450基因家族成员、BEE3转录因子、DIR1蛋白和MATE家族成员存在互作。CsERF025L启动子含有光响应、激素响应、应激响应等元件。基因CsERF025L在黄瓜种子时期表达量最高,显著高于叶、茎和根。比较CsERF025L在不同叶序黄瓜中的表达,发现对生叶序黄瓜cop1茎尖中CsERF025L表达量均显著高于互生叶序黄瓜R1。【结论】CsERF025L是一个位于细胞核的ERF转录 因子,可能在黄瓜对生叶序性状的形成与调控中具有潜在功能。
肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166.
XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber[J]. Biotechnology Bulletin, 2024, 40(4): 159-166.
| 用途 Application | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| 实时荧光定量PCR RT-qPCR | TUA-F | AGCGTTTGTCTGTTGACTATGGA |
| TUA-R | TGCAGATATCATAAATGGCTTCGT | |
| CsERF025L-qF | GGTTGCAATGCCGTTCTCAA | |
| CsERF025L-qR | ACAGCGCTTCTTCATCGACA | |
| 基因克隆 Gene cloning | CsERF025L-F | ATGGCCTCTACATCCTCCAAGAG |
| CsERF025L-R | CTAGTGGTAGCTCCAAAGATTTCCAT |
表1 CsERF025L转录因子实时荧光定量和克隆引物
Table 1 Primers for CsERF025L transcription factor cloning and real-time fluorescence quantification
| 用途 Application | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| 实时荧光定量PCR RT-qPCR | TUA-F | AGCGTTTGTCTGTTGACTATGGA |
| TUA-R | TGCAGATATCATAAATGGCTTCGT | |
| CsERF025L-qF | GGTTGCAATGCCGTTCTCAA | |
| CsERF025L-qR | ACAGCGCTTCTTCATCGACA | |
| 基因克隆 Gene cloning | CsERF025L-F | ATGGCCTCTACATCCTCCAAGAG |
| CsERF025L-R | CTAGTGGTAGCTCCAAAGATTTCCAT |

图1 黄瓜CsERF025L cDNA序列PCR扩增电泳图(A)和与参考序列比对结果(B)
Fig. 1 PCR amplification electrophoresis of cucumber CsERF025L cDNA sequences (A) and the aligned results with the reference sequence (B)
| 基因名称 Gene name | 氨基酸数 NAA | 分子质量 Mw/kD | 等电点 pI | 不稳定系数 II | 亲水系数 GRAVY | 带负电荷的残基总数 Asp+Glu | 带正电荷的残基总数 Arg+Lys | 脂肪族指数 AI | 亚细胞定位 SL |
|---|---|---|---|---|---|---|---|---|---|
| CsERF025L | 169 | 1.87 | 6.64 | 57.22 | -0.620 | 17 | 16 | 58.46 | 细胞核 |
表2 黄瓜CsERF025L蛋白理化性质分析
Table 2 Physicochemical properties of proteins in cucumber CsERF025L
| 基因名称 Gene name | 氨基酸数 NAA | 分子质量 Mw/kD | 等电点 pI | 不稳定系数 II | 亲水系数 GRAVY | 带负电荷的残基总数 Asp+Glu | 带正电荷的残基总数 Arg+Lys | 脂肪族指数 AI | 亚细胞定位 SL |
|---|---|---|---|---|---|---|---|---|---|
| CsERF025L | 169 | 1.87 | 6.64 | 57.22 | -0.620 | 17 | 16 | 58.46 | 细胞核 |

图2 黄瓜CsERF025L蛋白分析 A:黄瓜CsERF025L蛋白二级结构;B:黄瓜CsERF025L蛋白三级结构;C:黄瓜CsERF025L磷酸化预测;D:黄瓜CsERF025L蛋白互作网络模型图
Fig. 2 Protein analysis of cucumber CsERF025L A: Secondary structure of cucumber CsERF025L. B: Tertiary structure of cucumber CsERF025L protein. C: Prediction map of cucumber CsERF025L phosphorylation. D: Protein interaction network model of cucumber CsERF025L
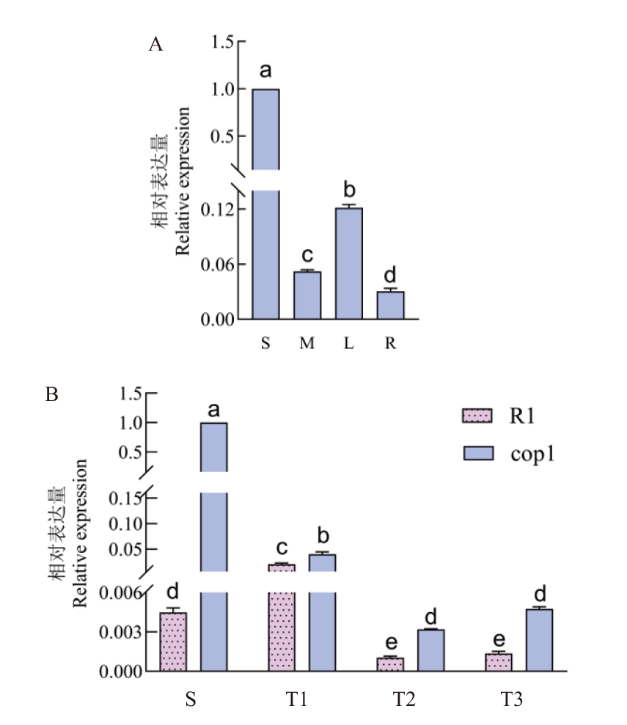
图6 CsERF025L在黄瓜中的特异性表达 S:种子(浸种后1 d);M:茎;L:叶;R:根;T1:茎尖1(浸种后7 d);T2:茎尖2(浸种后12 d);T3:茎尖3(浸种后17 d);R1:互生叶序黄瓜;cop1:对生叶序黄瓜;不同小写字母表示在0.05水平差异显著
Fig. 6 Specific expression of CsERF025L in cucumber S: Seed(the first day after soaking); M: stem; L: leaf; R: root; T1: stem tip 1(the seventh day after soaking); T2: stem tip 2(the 12th day after soaking; T3: stem tip 3(the 17th day after soaking); R1: alternate phyllotaxis in cucumber; cop1: opposite phyllotaxis in cucumber; Different lowercases indicate significant differences at the 0.05 level
| [4] |
Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors[J]. Biol Chem, 1998, 379(6): 633-646.
doi: 10.1515/bchm.1998.379.6.633 pmid: 9687012 |
| [5] | Gu C, Guo ZH, Hao PP, et al. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm[J]. Bot Stud, 2017, 58(1): 6. |
| [6] |
Gao Y, Han D, Jia W, et al. Molecular characterization and systematic analysis of NtAP2/ERF in tobacco and functional determination of NtRAV-4 under drought stress[J]. Plant Physiol Biochem, 2020, 156: 420-435.
doi: 10.1016/j.plaphy.2020.09.027 URL |
| [7] |
Nakano T, Suzuki K, Fujimura T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice[J]. Plant Physiol, 2006, 140(2): 411-432.
doi: 10.1104/pp.105.073783 URL |
| [8] |
潘健, 温海帆, 何欢乐, 等. 黄瓜ERF基因家族鉴定及其在雌花芽分化中的表达分析[J]. 中国农业科学, 2020, 53(1): 133-147.
doi: 10.3864/j.issn.0578-1752.2020.01.013 |
| Pan J, Wen HF, He HL, et al. Genome-wide identification of cucumber ERF gene family and expression analysis in female bud differentiation[J]. Sci Agric Sin, 2020, 53(1): 133-147. | |
| [9] |
Hu LF, Liu SQ. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers[J]. Genet Mol Biol, 2011, 34(4): 624-633.
doi: 10.1590/S1415-47572011005000054 URL |
| [10] | Sekhar S, Kumar J, Mohanty S, et al. Identification of novel QTLs for grain fertility and associated traits to decipher poor grain filling of basal spikelets in dense panicle rice[J]. Sci Rep, 2021, 11(1): 13617. |
| [11] | Xu Y, Li XN, Yang X, et al. Genome-wide identification and molecular characterization of the AP2/ERF superfamily members in sand pear(Pyrus pyrifolia)[J]. BMC Genomics, 2023, 24(1): 32. |
| [12] |
Lai PH, Huang LM, Pan ZJ, et al. PeERF1, a SHINE-like transcription factor, is involved in nanoridge development on lip epidermis of Phalaenopsis flowers[J]. Front Plant Sci, 2020, 10: 1709.
doi: 10.3389/fpls.2019.01709 URL |
| [13] |
Saelim L, Akiyoshi N, Tan TT, et al. Arabidopsis Group IIId ERF proteins positively regulate primary cell wall-type CESA genes[J]. J Plant Res, 2019, 132(1): 117-129.
doi: 10.1007/s10265-018-1074-1 |
| [14] |
Zhang M, Li ST, Nie L, et al. Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis[J]. Plant Mol Biol, 2015, 89(4-5): 463-473.
doi: 10.1007/s11103-015-0382-2 pmid: 26445975 |
| [15] | 崔馨文, 王燕燕. 喜树转录因子CaERF1原核表达及蛋白纯化[J/OL]. 分子植物育种, 2022. https://kns.cnki.net/kcms/detail/46.1068.S.20221015.1047.002.html. |
| Cui XW, Wang YY. Prokaryotic expression and purification of transcription factor CaERF1 from Camptotheca acuminata Decne[J/OL]. Molecular Plant Breeding, 2022. https://kns.cnki.net/kcms/detail/46.1068.S.20221015.1047.002.html. | |
| [16] | Yu F, Tan ZD, Fang T, et al. A comprehensive transcriptomics analysis reveals long non-coding RNA to be involved in the key metabolic pathway in response to waterlogging stress in maize[J]. Genes, 2020, 11(3): 267. |
| [17] | Park SI, Kwon HJ, et al. The OsERF115/AP2EREBP110 transcription factor is involved in the multiple stress tolerance to heat and drought in rice plants[J]. Int J Mol Sci, 2021, 22(13): 7181. |
| [18] | Xu Y, Lu JH, Zhang JD, et al. Transcriptome revealed the molecular mechanism of Glycyrrhiza inflata root to maintain growth and development, absorb and distribute ions under salt stress[J]. BMC Plant Biol, 2021, 21(1): 599. |
| [19] |
Sandhu D, Pudussery MV, et al. Characterization of natural genetic variation identifies multiple genes involved in salt tolerance in maize[J]. Funct Integr Genomics, 2020, 20(2): 261-275.
doi: 10.1007/s10142-019-00707-x |
| [20] |
Xie ZY, Wang J, Wang WS, et al. Integrated analysis of the transcriptome and metabolome revealed the molecular mechanisms underlying the enhanced salt tolerance of rice due to the application of exogenous melatonin[J]. Front Plant Sci, 2021, 11: 618680.
doi: 10.3389/fpls.2020.618680 URL |
| [21] | 徐乐, 陈培茹, 刘意, 等. 甘薯IbERF071基因的克隆及表达特性分析[J]. 长江大学学报: 自然科学版, 2022, 19(3): 99-108. |
| Xu L, Chen PR, Liu Y, et al. Cloning and expression pattern analysis of IbERF071 gene from sweet potato[J]. J Yangtze Univ Nat Sci Ed, 2022, 19(3): 99-108. | |
| [22] |
Tahmasebi A, Khahani B, Tavakol E, et al. Microarray analysis of Arabidopsis thaliana exposed to single and mixed infections with Cucumber mosaic virus and turnip viruses[J]. Physiol Mol Biol Plants, 2021, 27(1): 11-27.
doi: 10.1007/s12298-021-00925-3 |
| [23] |
Tezuka D, Kawamata A, et al. The rice ethylene response factor OsERF83 positively regulates disease resistance to Magnaporthe oryzae[J]. Plant Physiol Biochem, 2019, 135: 263-271.
doi: 10.1016/j.plaphy.2018.12.017 URL |
| [24] |
Gebretsadik K, Qiu XY, Dong SY, et al. Molecular research progress and improvement approach of fruit quality traits in cucumber[J]. Theor Appl Genet, 2021, 134(11): 3535-3552.
doi: 10.1007/s00122-021-03895-y pmid: 34181057 |
| [25] | Gou CX, Zhu PY, Meng YJ, et al. Evaluation and genetic analysis of parthenocarpic germplasms in cucumber[J]. Genes, 2022, 13(2): 225. |
| [26] | 刘坤. 中间锦鸡儿瞬时表达体系的建立及AP2/ERF基因家族的鉴定、分析和功能研究[D]. 呼和浩特: 内蒙古农业大学, 2018. |
| Liu K. Establishment of transient expression system and identification, analysis and functional study of AP2/ERF gene family from Caragana intermedia[D]. Hohhot: Inner Mongolia Agricultural University, 2018. | |
| [27] | Tang YS, Zhang GR, Jiang XY, et al. Genome-wide association study of glucosinolate metabolites(mGWAS)in Brassica napus L[J]. Plants, 2023, 12(3): 639. |
| [28] |
Wang CH, Xin M, Zhou XY, et al. The novel ethylene-responsive factor CsERF025 affects the development of fruit bending in cucumber[J]. Plant Mol Biol, 2017, 95(4-5): 519-531.
doi: 10.1007/s11103-017-0671-z pmid: 29052099 |
| [29] | Mann R, Notani D. Transcription factor condensates and signaling driven transcription[J]. Nucleus, 2023, 14(1): 2205758. |
| [30] | 张通. bHLH类转录因子RhBEE3通过促进ABA合成调节月季花朵衰老的机理研究[D]. 武汉: 华中农业大学, 2021. |
| Zhang T. The mechanism of bHLH transcription factor RhBEE3 mediates rose flower senescence by promoting ABA synthesis[D]. Wuhan: Huazhong Agricultural University, 2021. | |
| [31] | 肖亮. AP2/ERF转录因子LTF1调控菘蓝活性木脂素生物合成的机制研究[D]. 上海: 中国人民解放军海军军医大学, 2021. |
| Xiao L. Study on the mechanism of AP2/ERF transcription factor LTF1 regulating lignan biosynthesis in Isatis indigotica[D]. Shanghai: Naval Medical University, 2021. | |
| [32] | 李瑞熙, 李洁如, 瞿礼嘉. 拟南芥MATE家族基因ADP1通过调节生长素水平调控植物株型建成[C]// 2011全国植物生物学研讨会论文集. 南宁, 2011: 33. |
| Li RX, Li JR, Qu LJ. Arabidopsis MATE family gene ADP1 regulates plant formation by regulating auxin levels[C]// 2011Proceedings of the National Symposium on Plant Biology. Nanning, 2011: 33. | |
| [33] |
Scheres B, Krizek BA. Coordination of growth in root and shoot apices by AIL/PLT transcription factors[J]. Curr Opin Plant Biol, 2018, 41: 95-101.
doi: S1369-5266(17)30175-9 pmid: 29121612 |
| [34] |
Würschum T, Gross-Hardt R, Laux T. APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem[J]. Plant Cell, 2006, 18(2): 295-307.
doi: 10.1105/tpc.105.038398 pmid: 16387832 |
| [35] |
Lee DY, Lee J, Moon S, et al. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem[J]. Plant J, 2007, 49(1): 64-78.
doi: 10.1111/tpj.2007.49.issue-1 URL |
| [1] |
Xie W, Ding CQ, Hu HT, et al. Molecular events of rice AP2/ERF transcription factors[J]. Int J Mol Sci, 2022, 23(19): 12013.
doi: 10.3390/ijms231912013 URL |
| [2] |
Sharoni AM, Nuruzzaman M, Satoh K, et al. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice[J]. Plant Cell Physiol, 2011, 52(2): 344-360.
doi: 10.1093/pcp/pcq196 pmid: 21169347 |
| [3] |
Allen MD, Yamasaki K, Ohme-Takagi M, et al. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA[J]. EMBO J, 1998, 17(18): 5484-5496.
doi: 10.1093/emboj/17.18.5484 pmid: 9736626 |
| [1] | 娄银, 高浩竣, 王茜, 牛景萍, 王敏, 杜维俊, 岳爱琴. 大豆GmHMGS基因的鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(4): 110-121. |
| [2] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [3] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [4] | 张清兰, 张亚冉, 鞠志花, 王秀革, 肖遥, 王金鹏, 魏晓超, 高亚平, 白福恒, 王洪程. 牛TARDBP基因核心启动子鉴定与转录调控分析[J]. 生物技术通报, 2024, 40(4): 306-318. |
| [5] | 陈晓松, 刘超杰, 郑佳, 乔宗伟, 罗惠波, 邹伟. TMT定量蛋白质组学解析Rummeliibacillus suwonensis 3B-1 生长及己酸代谢机制[J]. 生物技术通报, 2024, 40(3): 135-145. |
| [6] | 杨伟成, 孙岩, 杨倩, 王壮琳, 马菊花, 薛金爱, 李润植. 陆地棉FAX家族的全基因组鉴定及GhFAX1的功能分析[J]. 生物技术通报, 2024, 40(3): 155-169. |
| [7] | 杨伟杰, 杨周林, 朱浩东, 魏煜, 刘君, 刘训. 地衣素合成酶关键模块 LchAD 蛋白的性质和功能研究[J]. 生物技术通报, 2024, 40(3): 322-332. |
| [8] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [9] | 龚丽丽, 余花, 杨杰, 陈天池, 赵双滢, 吴月燕. 葡萄CYP707A基因家族的鉴定及对果实成熟的功能验证[J]. 生物技术通报, 2024, 40(2): 160-171. |
| [10] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [11] | 谢宏, 周丽莹, 李舒文, 王梦迪, 艾晔, 晁跃辉. 蒺藜苜蓿MtCIM基因结构和功能分析[J]. 生物技术通报, 2024, 40(1): 262-269. |
| [12] | 唐伟林, 康琴, 汪霞, 谌明洋, 孙欣江, 王棵, 侯凯, 吴卫, 徐东北. 薄荷茉莉酸受体McCOI1a基因的克隆与表达模式分析[J]. 生物技术通报, 2024, 40(1): 270-280. |
| [13] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [14] | 褚睿, 李昭轩, 张学青, 杨东亚, 曹行行, 张雪艳. 黄瓜枯萎病拮抗芽孢杆菌的筛选、鉴定及其生防潜力[J]. 生物技术通报, 2023, 39(8): 262-271. |
| [15] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||