








生物技术通报 ›› 2024, Vol. 40 ›› Issue (8): 53-62.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0058
孙志勇1( ), 杜怀东1, 刘阳1, 马嘉欣1, 于雪然1,2, 马伟1, 姚鑫杰1, 王敏1, 李培富1(
), 杜怀东1, 刘阳1, 马嘉欣1, 于雪然1,2, 马伟1, 姚鑫杰1, 王敏1, 李培富1( )
)
收稿日期:2024-01-15
出版日期:2024-08-26
发布日期:2024-09-05
通讯作者:
李培富,男,博士,教授,研究方向:水稻遗传育种;E-mail: peifuli@163.com作者简介:孙志勇,男,硕士研究生,研究方向:水稻功能品质遗传育种;E-mail: 1055196309@qq.com
基金资助:
SUN Zhi-yong1( ), DU Huai-dong1, LIU Yang1, MA Jia-xin1, YU Xue-ran1,2, MA Wei1, YAO Xin-jie1, WANG Min1, LI Pei-fu1(
), DU Huai-dong1, LIU Yang1, MA Jia-xin1, YU Xue-ran1,2, MA Wei1, YAO Xin-jie1, WANG Min1, LI Pei-fu1( )
)
Received:2024-01-15
Published:2024-08-26
Online:2024-09-05
摘要:
【目的】挖掘与水稻籽粒γ-氨基丁酸含量显著相关的SNP位点及候选基因,为揭示水稻籽粒γ-氨基丁酸合成与积累的遗传基础和分子机制提供理论依据,为高γ-氨基丁酸含量的水稻品种选育奠定基础。【方法】以139份西北早粳稻核心种质为实验材料,测定其籽粒γ-氨基丁酸含量,结合重测序基因型数据,进行全基因组关联(GWAS)分析,根据单倍型分析和基因注释筛选候选基因,通过表达谱数据库和荧光定量 PCR 对预测基因进行表达模式分析。【结果】水稻籽粒γ-氨基丁酸含量表现出丰富的变异,变异系数为31.25%。全基因组关联分析检测到分别位于第6、8、9、11和12号染色体上共6个有代表性的SNP位点,筛选出已克隆与γ-氨基丁酸含量有关的OsBADH2和OsGABA-T2基因,预测出6个与γ-氨基丁酸含量有关的候选基因,将LOC_Os09g10720基因列为重点分析基因,并进一步验证了其参与水稻籽粒中GABA分解的可能性。【结论】检测到6个与水稻籽粒γ-氨基丁酸含量显著关联的 SNP 位点。筛出6个可能与水稻籽粒γ-氨基丁酸含量相关的候选基因,分别是LOC_Os09g09750、LOC_Os09g09760、LOC_Os09g10210、LOC_Os09g09820、LOC_Os09g10280和LOC_Os09g10720,其中LOC_Os09g10720作为重点候选基因。
孙志勇, 杜怀东, 刘阳, 马嘉欣, 于雪然, 马伟, 姚鑫杰, 王敏, 李培富. 水稻籽粒γ-氨基丁酸含量的全基因组关联分析[J]. 生物技术通报, 2024, 40(8): 53-62.
SUN Zhi-yong, DU Huai-dong, LIU Yang, MA Jia-xin, YU Xue-ran, MA Wei, YAO Xin-jie, WANG Min, LI Pei-fu. Genome-wide Association Analysis of γ-aminobutyric Acid in Rice Grains[J]. Biotechnology Bulletin, 2024, 40(8): 53-62.
| 变异来源Source of variation | 自由度Degree of freedom | 平方和Sum of squares | 均方Mean square | F值F value | P 值P value |
|---|---|---|---|---|---|
| 群组间Between-group | 138 | 133 214.515 | 965.323 | 213.578 | 0.000 |
| 群组内Within-group | 278 | 1 256.493 | 4.520 | ||
| 总计Total | 416 | 134 471.008 |
表1 139份种质资源GABA含量方差分析
Table 1 Variance analysis of GABA content in 139 varieties of germplasm resources
| 变异来源Source of variation | 自由度Degree of freedom | 平方和Sum of squares | 均方Mean square | F值F value | P 值P value |
|---|---|---|---|---|---|
| 群组间Between-group | 138 | 133 214.515 | 965.323 | 213.578 | 0.000 |
| 群组内Within-group | 278 | 1 256.493 | 4.520 | ||
| 总计Total | 416 | 134 471.008 |
| SNP | 染色体 Chromosome | 位置 Location/bp | 置信区间起点 Starting point of confidence interval | 置信区间终点 End point of confidence interval | 参考碱基 Reference base | 变异碱基 Variant base | -log10(P) |
|---|---|---|---|---|---|---|---|
| S6-3357208 | 6 | 3357208 | 3057208 | 3657208 | G | A | 7.07 |
| S8-6442742 | 8 | 6442742 | 6142742 | 6742742 | A | C | 6.51 |
| S8-20231522 | 8 | 20231522 | 19931522 | 20531522 | T | C | 7.23 |
| S9-5559515 | 9 | 5559515 | 5259515 | 5859515 | C | T | 6.45 |
| S11-4162881 | 11 | 4162881 | 3862881 | 4462881 | C | T | 6.80 |
| S12-12473570 | 12 | 12473570 | 12173570 | 12773570 | T | C | 6.26 |
表2 全基因组关联分析显著SNPs位点
Table 2 Genome-wide association analysis of significant SNPs loci
| SNP | 染色体 Chromosome | 位置 Location/bp | 置信区间起点 Starting point of confidence interval | 置信区间终点 End point of confidence interval | 参考碱基 Reference base | 变异碱基 Variant base | -log10(P) |
|---|---|---|---|---|---|---|---|
| S6-3357208 | 6 | 3357208 | 3057208 | 3657208 | G | A | 7.07 |
| S8-6442742 | 8 | 6442742 | 6142742 | 6742742 | A | C | 6.51 |
| S8-20231522 | 8 | 20231522 | 19931522 | 20531522 | T | C | 7.23 |
| S9-5559515 | 9 | 5559515 | 5259515 | 5859515 | C | T | 6.45 |
| S11-4162881 | 11 | 4162881 | 3862881 | 4462881 | C | T | 6.80 |
| S12-12473570 | 12 | 12473570 | 12173570 | 12773570 | T | C | 6.26 |

图3 SNP S9-5559515的LD热图(A)及候选基因的单倍型分析(B-G) a、b 表示 0.05 水平差异
Fig. 3 LD heatmap of SNP S9-5559515 (A) and haplotype analysis of candidate genes (B-G) a and b indicate significant differences at the 0.05 level
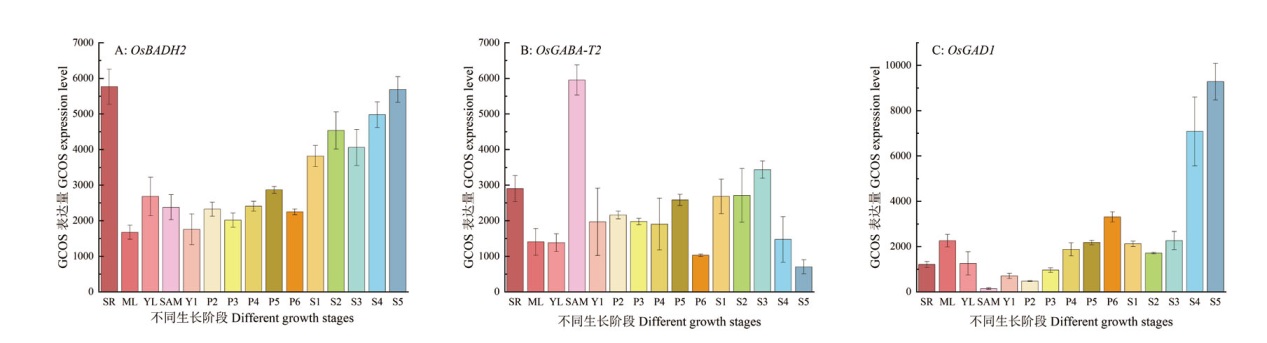
图4 水稻籽粒GABA含量关键基因表达图谱 SR:胚根;ML:成熟叶;YL:幼叶;SAM:顶端分生组织和轴分生组织的萌发期;YI:花过渡与花器官发育期;P2-P3:减数分裂阶段;P4:幼年小孢子期;P5:液泡化花粉期;P6:成熟花粉期;S1:球形胚早期;S2:球形胚中晚期;S3:种子形态发生期;S4:种子成熟期;S5:种子耐休眠和脱水期,下同
Fig. 4 Expression patterns of key genes for GABA content in rice grains SR: Radicle. ML: Mature leaf.YL: Young leaf. SAM: Germination stage of apical meristem and axial meristem. YI: Flower transition and floral organ development stage. P2-P3: Meiosis stage. P4: Young microspore stage. P5: Vacuolated pollen stage. P6: Mature pollen stage. S1: Early globular embryo stage. S2: Mid-late globular embryo stage. S3: Seed morphogenesis stage. S4: Seed maturity stage.S5: Seed dormancy and dehydration tolerance stage, the same below
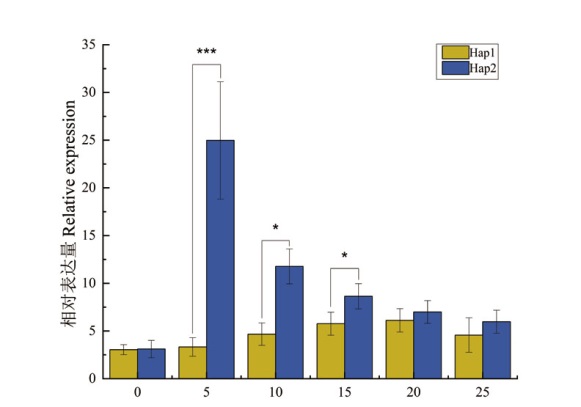
图6 LOC_Os09g10720基因在花后不同时期相对表达水平 * 与 *** 分别表示0.05 和0.001 的显著水平
Fig. 6 Relative expressions of LOC_Os09g10720 genes at different stages after flowering * and *** indicates significant at 0.05 and 0.001 probability levels respectively
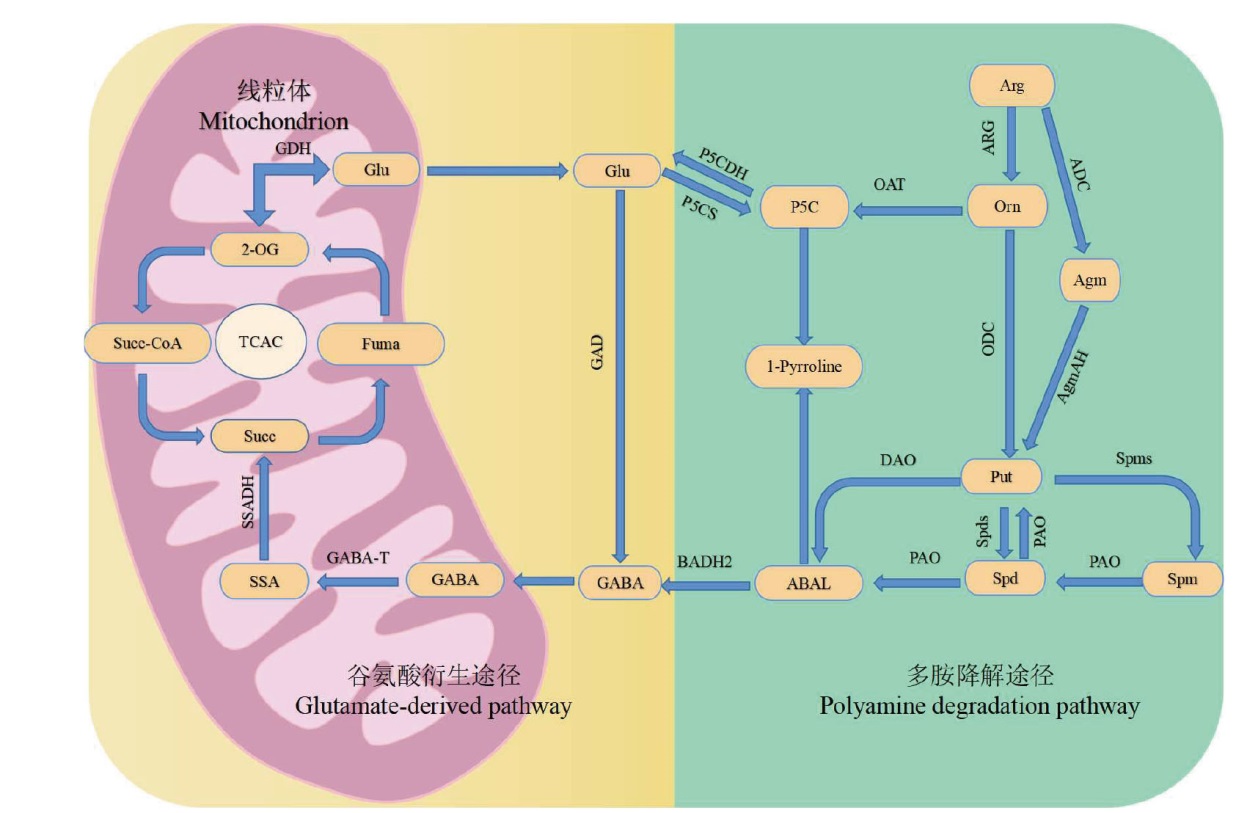
图7 GABA代谢通路 Glu:谷氨酸;2-OG:α-酮戊二酸;Succ-CoA: 琥珀酰辅酶A;Fuma: 延胡索酸;Succ: 琥珀酸;SSA: 琥珀酸半醛;Arg: 精氨酸;P5C: 1-吡咯啉-5-羧酸;Orn: 鸟氨酸;Agm: 鲱精胺;1-Pyrroline: 1-吡咯啉;Put: 腐胺;ABAL: 4-氨基丁醛;Spd: 亚精胺;Spm: 精胺;GDH: 谷氨酸脱氢酶;SSADH: 琥珀酸半醛脱氢酶;GABA-T:γ- 氨基丁酸转氨酶;GAD:谷氨酸脱羧酶;P5CDH:吡咯啉-5-羧酸脱氢酶:P5CS: 1-吡咯啉-5-羧酸合成酶;BADH2:甜菜碱醛脱氢酶2;OAT:鸟氨酸转氨酶;ARG:精氨酸酶;ADC: 精氨酸脱羧酶;ODC:鸟氨酸脱羧酶;AgmAH:鲱精胺酶;DAO: 二胺氧化酶;Spms: 精胺合成酶;PAO:多胺氧化酶;SpdS:亚精胺合成酶
Fig. 7 GABA metabolic pathway Glu: Glutamic acid; 2-OG: α-ketoglutaric acid; Succ-CoA: Succinyl-CoA; Fuma: Fumaric acid; Succ: Succinic acid; SSA: Succinic semialdehyde; Arg: Arginine; P5C: 1-pyrroline-5-carboxylic acid; Orn: Ornithine; Agm: Agmatine; 1-pyrroline: 1-pyrroline; Put: Putrescine; ABAL: 4-aminobutyraldehyde; Spd: Spermidine; Spm: Spermine; GDH: Glutamate dehydrogenase; SSADH: Succinic semialdehyde dehydrogenase; GABA-T: γ-aminobutyric acid transaminase; GAD: Glutamic acid decarboxylase; P5CDH: Pyrroline-5-carboxylic acid dehydrogenase; P5CS: 1-pyrroline-5-carboxylic acid synthetase; BADH2: Betaine aldehyde dehydrogenase 2; OAT: ornithine aminotransferase; ARG: Arginase; ADC: Arginine decarboxylase; ODC: Ornithine decarboxylase; AgmAH: Agmatine amidohydrolase; DAO: Diamine oxidase; Spms: Spermine synthetase; PAO: Polyamine oxidase; SpdS: Spermidine synthetase
| [1] |
Inoue K, Shirai T, Ochiai H, et al. Blood-pressure-lowering effect of a novel fermented milk containing gamma-aminobutyric acid(GABA)in mild hypertensives[J]. Eur J Clin Nutr, 2003, 57(3): 490-495.
doi: 10.1038/sj.ejcn.1601555 pmid: 12627188 |
| [2] | Schweizer-Schubert S, Gordon JL, Eisenlohr-Moul TA, et al. Steroid hormone sensitivity in reproductive mood disorders: on the role of the GABAA receptor complex and stress during hormonal transitions[J]. Front Med, 2021, 7: 479646. |
| [3] | Oketch-Rabah HA, Madden EF, Roe AL, et al. United states pharmacopeia(USP)safety review of Gamma-aminobutyric acid(GABA)[J]. Nutrients, 2021, 13(8): 2742. |
| [4] | Gramazio P, Takayama M, Ezura H. Challenges and prospects of new plant breeding techniques for GABA improvement in crops: tomato as an example[J]. Front Plant Sci, 2020, 11: 577980. |
| [5] | Yu YJ, Li M, Li CX, et al. Accelerated accumulation of γ-aminobutyric acid and modifications on its metabolic pathways in black rice grains by germination under cold stress[J]. Foods, 2023, 12(6): 1290. |
| [6] | Shelp BJ, Zarei A. Subcellular compartmentation of 4-aminobutyrate(GABA)metabolism in Arabidopsis: an update[J]. Plant Signal Behav, 2017, 12(5): e1322244. |
| [7] | 黄志伟, 许明, 林忠辉, 等. 籼稻GAD基因的克隆、序列分析及其植物表达载体构建[J]. 南方农业学报, 2012, 43(8): 1079-1085. |
| Huang ZW, Xu M, Lin ZH, et al. Cloning, sequence analysis of GAD gene from indica rice and its plant expression vector construction[J]. J South Agric, 2012, 43(8): 1079-1085. | |
| [8] | 向文豪, 崔国鹏, 王圣洁, 等. 基于Diurnal在线软件分析拟南芥生物节律基因对GABA的响应[J]. 安徽农学通报, 2014, 20(8): 24-27, 45. |
| Xiang WH, Cui GP, Wang SJ, et al. Analyzing gene expression of circadian clock in responding to GABA in Arabidopsis based on diurnal-online software application[J]. Anhui Agric Sci Bull, 2014, 20(8): 24-27, 45. | |
| [9] | 李敬蕊, 田真, 吴晓蕾, 等. 小白菜GAD基因克隆及高氮条件下外源GABA的诱导表达分析[J]. 农业生物技术学报, 2017, 25(8): 1217-1227. |
| Li JR, Tian Z, Wu XL, et al. Cloning of GAD gene in pakchoi(Brassica campestris ssp. chinensis)and induced expression analysis treated with exogenous GABA under higher nitrogen level[J]. J Agric Biotechnol, 2017, 25(8): 1217-1227. | |
| [10] |
李明轩, 刘颖, 杨柏明, 等. 西瓜谷氨酸脱羧酶GADs的克隆与表达分析[J]. 华北农学报, 2023, 38(6): 18-25.
doi: 10.7668/hbnxb.20194340 |
| Li MX, Liu Y, Yang BM, et al. Cloning and expression analysis of glutamate decarboxylase GADs from watermelon[J]. Acta Agric Boreali Sin, 2023, 38(6): 18-25. | |
| [11] | Akama K, Akter N, Endo H, et al. An in vivo targeted deletion of the calmodulin-binding domain from rice glutamate decarboxylase 3(OsGAD3)increases γ-aminobutyric acid content in grains[J]. Rice, 2020, 13(1): 20. |
| [12] | Agudelo-Romero P, Bortolloti C, Pais MS, et al. Study of polyamines during grape ripening indicate an important role of polyamine catabolism[J]. Plant Physiol Biochem, 2013, 67: 105-119. |
| [13] | Liao YX, Li MY, Wu HZ, et al. Generation of aroma in three-line hybrid rice through CRISPR/Cas9 editing of BETAINE ALDEHYDE DEHYDROGENASE2(OsBADH2)[J]. Physiol Plant, 2024, 176(1): e14206. |
| [14] | Shimajiri Y, Ozaki K, Kainou K, et al. Differential subcellular localization, enzymatic properties and expression patterns of γ-aminobutyric acid transaminases(GABA-Ts)in rice(Oryza sativa)[J]. J Plant Physiol, 2013, 170(2): 196-201. |
| [15] | Tola AJ, Jaballi A, Germain H, et al. Recent development on plant aldehyde dehydrogenase enzymes and their functions in plant development and stress signaling[J]. Genes, 2020, 12(1): 51. |
| [16] | Hidalgo-Castellanos J, Duque AS, Burgueño A, et al. Overexpression of the arginine decarboxylase gene promotes the symbiotic interaction Medicago truncatula-Sinorhizobium meliloti and induces the accumulation of proline and spermine in nodules under salt stress conditions[J]. J Plant Physiol, 2019, 241: 153034. |
| [17] | Bukomarhe CB, Kimwemwe PK, Githiri SM, et al. Association mapping of candidate genes associated with iron and zinc content in rice(Oryza sativa L.)grains[J]. Genes, 2023, 14(9): 1815. |
| [18] |
王龙海, 杨泽伟, 朱莉, 等. 甜高粱琥珀酸半醛脱氢酶SbSSADH基因克隆及原核表达[J]. 生物技术通报, 2015, 31(7): 83-90.
doi: 10.13560/j.cnki.biotech.bull.1985.2015.07.013 |
| Wang LH, Yang ZW, Zhu L, et al. Cloning and prokaryotic expression of sweet sorghum succinic semialdehyde dehydrogenase SbSSADH[J]. Biotechnol Bull, 2015, 31(7): 83-90. | |
| [19] | Alqudah AM, Sallam A, Stephen Baenziger P, et al. GWAS: fast-forwarding gene identification and characterization in temperate Cereals: lessons from Barley - A review[J]. J Adv Res, 2019, 22: 119-135. |
| [20] | Ntakirutimana F, Tranchant-Dubreuil C, Cubry P, et al. Genome-wide association analysis identifies natural allelic variants associated with panicle architecture variation in African rice, Oryza glaberrima Steud[J]. G3, 2023, 13(10): jkad174. |
| [21] |
刘忠奇, 张海清, 贺记外, 等. 成熟期水稻种子脱水速率全基因组关联分析[J]. 中国水稻科学, 2024, 38(2): 150-159.
doi: 10.16819/j.1001-7216.2024.230305 |
|
Liu ZQ, Zhang HQ, He JW, et al. Genome-wide association analysis of rice seed dehydration rate at maturity stage[J]. Chin J Rice Sci, 2024, 38(2): 150-159.
doi: 10.16819/j.1001-7216.2024.230305 |
|
| [22] | 孙晓雪. 水稻功能性营养品质性状QTL定位及环境互作分析[D]. 哈尔滨: 东北农业大学, 2017. |
| Sun XX. Analysis of qtl mapping and environmental interaction of functional nutritional quality traits in rice[D]. Harbin: Northeast Agricultural University, 2017. | |
| [23] | 田玲, 王康恺, 王迎超, 等. 水稻籽粒γ-氨基丁酸含量的QTL定位分析[J]. 植物遗传资源学报, 2019, 20(6): 1517-1522. |
| Tian L, Wang KK, Wang YC, et al. QTL mapping for the genetic components determining the rice grain γ-aminobutyric acid content[J]. J Plant Genet Resour, 2019, 20(6): 1517-1522. | |
| [24] | 王迎超, 王全兴, 王浩, 等. 富γ-氨基丁酸水稻种质筛选及与子粒性状相关性分析[J]. 植物遗传资源学报, 2016, 17(6): 1116-1122. |
| Wang YC, Wang QX, Wang H, et al. Selection of rice germplasm with rich γ-aminobutyric acid and correlation analysis of the content and grain traits[J]. J Plant Genet Resour, 2016, 17(6): 1116-1122. | |
| [25] | 陈雪, 刘明, 汪丽萍, 等. 柱前在线衍生-HPLC法测定发芽糙米中γ-氨基丁酸含量[J]. 粮油食品科技, 2017, 25(3): 48-53. |
| Chen X, Liu M, Wang LP, et al. Determination of γ-aminobutyric acid in germinated brown rice by online pre-column derivatization combined with HPLC[J]. Sci Technol Cereals Oils Foods, 2017, 25(3): 48-53. | |
| [26] | Hui SZ, Li HJ, Mawia AM, et al. Production of aromatic three-line hybrid rice using novel alleles of BADH2[J]. Plant Biotechnol J, 2022, 20(1): 59-74. |
| [27] | 周露, 沈贝贝, 白苏阳, 等. 以RNA干扰γ-氨基丁酸转氨酶1基因(OsGABA-T1)表达提高稻米γ-氨基丁酸(GABA)含量[J]. 作物学报, 2015, 41(9): 1305-1312. |
| Zhou L, Shen BB, Bai SY, et al. RNA interference of Os GABA-T1 gene expression induced GABA accumulation in rice grain[J]. Acta Agron Sin, 2015, 41(9): 1305-1312. | |
| [28] |
潘阳阳, 陈宜波, 王重荣, 等. γ-氨基丁酸和2-乙酰-1-吡咯啉代谢通路在水稻籽粒发育过程中的变化分析[J]. 中国水稻科学, 2021, 35(2): 121-129.
doi: 10.16819/j.1001-7216.2021.0805 |
| Pan YY, Chen YB, Wang CR, et al. Metabolism of γ-aminobutyrate and 2-acetyl-1-pyrroline analyses at various grain developmental stages in rice(Oryza sativa L.)[J]. Chin J Rice Sci, 2021, 35(2): 121-129. | |
| [29] | 胡伟, 杨晓颖, 李美英, 等. 香蕉谷氨酸脱羧酶基因克隆与表达[J]. 西北植物学报, 2009, 29(3): 429-434. |
| Hu W, Yang XY, Li MY, et al. Cloning and expression analysis of MaGAD1 gene from banana fruit[J]. Acta Bot Boreali Occidentalia Sin, 2009, 29(3): 429-434. | |
| [30] | Michaeli S, Fait A, Lagor K, et al. A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism[J]. Plant J, 2011, 67(3): 485-498. |
| [31] | Podlešáková K, Ugena L, Spíchal L, et al. Phytohormones and polyamines regulate plant stress responses by altering GABA pathway[J]. N Biotechnol, 2019, 48: 53-65. |
| [1] | 李庆懋, 彭聪归, 齐笑含, 刘兴蕾, 李臻园, 李沁妍, 黄立钰. 促进水稻铁素吸收的野生稻内生细菌优良菌株的筛选与鉴定[J]. 生物技术通报, 2024, 40(8): 255-263. |
| [2] | 庞梦真, 徐汉琴, 刘海燕, 宋娟, 王佳涵, 孙丽娜, 姬佩梅, 尹泽芝, 胡又川, 赵晓萌, 梁闪闪, 张泗举, 栾维江. 水稻黄化早抽穗突变体 hz1 的基因鉴定及功能分析[J]. 生物技术通报, 2024, 40(7): 125-136. |
| [3] | 田胜尼, 张琴, 董玉飞, 丁洲, 叶爱华, 张明珠. 酸性矿山废水对成熟期水稻根区理化因子及固氮微生物的影响[J]. 生物技术通报, 2024, 40(6): 271-280. |
| [4] | 孔小平, 陈利文, 刘思思, 严湘萍. 胡萝卜抽薹相关性状全基因组关联分析[J]. 生物技术通报, 2024, 40(5): 120-130. |
| [5] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [6] | 李兴容, 谭志兵, 赵燕, 李曜魁, 赵炳然, 唐丽. 水稻低亲和性阳离子转运蛋白基因OsLCT3的克隆与功能研究[J]. 生物技术通报, 2024, 40(4): 97-109. |
| [7] | 单新雨, 李太春, 杨若晨, 段香茹, 康佳, 张英杰, 刘月琴. γ-氨基丁酸对绵羊卵巢颗粒细胞凋亡及类固醇激素分泌的影响[J]. 生物技术通报, 2024, 40(3): 312-321. |
| [8] | 李雪, 李容欧, 孔美懿, 黄磊. 解淀粉芽孢杆菌SQ-2对水稻的促生作用[J]. 生物技术通报, 2024, 40(2): 109-119. |
| [9] | 邹修为, 岳佳妮, 李志宇, 戴良英, 李魏. 水稻热激转录因子HsfA2b调控非生物胁迫抗性的功能分析[J]. 生物技术通报, 2024, 40(2): 90-98. |
| [10] | 张超, 王子瑞, 孙亚丽, 毛馨晨, 唐家琪, 于恒秀. 水稻维生素B1合成相关基因OsTHIC的功能研究[J]. 生物技术通报, 2024, 40(2): 99-108. |
| [11] | 周会汶, 吴兰花, 韩德鹏, 郑伟, 余跑兰, 吴杨, 肖小军. 甘蓝型油菜种子硫苷含量全基因组关联分析[J]. 生物技术通报, 2024, 40(1): 222-230. |
| [12] | 林鑫焱, 张传忠, 戴兵, 王馨珩, 刘剑锋, 温丽, 徐兴健, 方军. 水稻穗发芽遗传与分子机制的研究进展[J]. 生物技术通报, 2024, 40(1): 24-31. |
| [13] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [14] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [15] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||