








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 125-136.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0132
庞梦真1,2( ), 徐汉琴1,2, 刘海燕1,2, 宋娟1,2, 王佳涵1,2, 孙丽娜1,2, 姬佩梅1,2, 尹泽芝1,2, 胡又川1,2, 赵晓萌1,2, 梁闪闪1,2, 张泗举1,2(
), 徐汉琴1,2, 刘海燕1,2, 宋娟1,2, 王佳涵1,2, 孙丽娜1,2, 姬佩梅1,2, 尹泽芝1,2, 胡又川1,2, 赵晓萌1,2, 梁闪闪1,2, 张泗举1,2( ), 栾维江1,2(
), 栾维江1,2( )
)
收稿日期:2024-02-02
出版日期:2024-07-26
发布日期:2024-05-24
通讯作者:
栾维江,男,博士,教授,研究方向:水稻功能基因组学;E-mail: lwjzsq@163.com;作者简介:庞梦真,女,硕士研究生,研究方向:遗传学;E-mail: mengzhenpang@163.com
基金资助:
PANG Meng-zhen1,2( ), XU Han-qin1,2, LIU Hai-yan1,2, SONG Juan1,2, WANG Jia-han1,2, SUN Li-na1,2, JI Pei-mei1,2, YIN Ze-zhi1,2, HU You-chuan1,2, ZHAO Xiao-meng1,2, LIANG Shan-shan1,2, ZHANG Si-ju1,2(
), XU Han-qin1,2, LIU Hai-yan1,2, SONG Juan1,2, WANG Jia-han1,2, SUN Li-na1,2, JI Pei-mei1,2, YIN Ze-zhi1,2, HU You-chuan1,2, ZHAO Xiao-meng1,2, LIANG Shan-shan1,2, ZHANG Si-ju1,2( ), LUAN Wei-jiang1,2(
), LUAN Wei-jiang1,2( )
)
Received:2024-02-02
Published:2024-07-26
Online:2024-05-24
摘要:
【目的】 水稻的抽穗期对水稻地域适应性及水稻产量至关重要,对水稻抽穗期基因进行鉴定及功能分析,可以为水稻育种提供优异的基因资源。【方法】 通过BSA-seq方法对一个黄化早抽穗突变体hz1(huangzao 1)进行基因定位克隆及连锁分析;利用RT-qPCR技术分析目的基因HZ1的表达谱,并用水稻原生质体瞬时转化查明HZ1的亚细胞定位;对突变体的抽穗期、叶绿素含量、过氧化氢含量等生理指标进行测定,详细分析其表型变化。【结果】 田间表型观察发现hz1表现早抽穗,长日照(long-day, LD)及短日照(short-day, SD)条件下hz1的抽穗期相同,分别比野生型(wild type, WT)早抽穗43 d和26 d,表明hz1是一个光周期不敏感的突变体。同时hz1表现黄化表型,相比WT,叶绿素含量下降。遗传分析表明hz1由隐性单基因控制,F2混池高通量测序将目的基因定位于水稻第6染色体上17.8 Mb区间内,分析发现该区间内一个T-DNA插入位点LOC_Os06g40080与hz1目标性状完全连锁,LOC_Os06g40080为已知的SE5基因,编码血红素加氧酶1(heme oxygenase 1, HO1)。HZ1/SE5在叶片中高表达,在LD及SD条件下具有昼夜节律性表达。亚细胞定位发现HZ1/SE5蛋白定位于叶绿体中。表达调控分析表明HZ1/SE5主要通过调控水稻成花素Hd3a和RFT1的表达来调控水稻的抽穗期;并通过调控叶绿素合成途径相关基因的表达水平影响水稻叶绿素水平变化。【结论】 黄化早抽穗突变体hz1由于血红素加氧酶编码基因SE5突变导致其对光周期不敏感,HZ1/SE5基因通过调控水稻成花素基因及叶绿素合成途径相关基因的表达而影响水稻的抽穗期及叶片的黄化。
庞梦真, 徐汉琴, 刘海燕, 宋娟, 王佳涵, 孙丽娜, 姬佩梅, 尹泽芝, 胡又川, 赵晓萌, 梁闪闪, 张泗举, 栾维江. 水稻黄化早抽穗突变体 hz1 的基因鉴定及功能分析[J]. 生物技术通报, 2024, 40(7): 125-136.
PANG Meng-zhen, XU Han-qin, LIU Hai-yan, SONG Juan, WANG Jia-han, SUN Li-na, JI Pei-mei, YIN Ze-zhi, HU You-chuan, ZHAO Xiao-meng, LIANG Shan-shan, ZHANG Si-ju, LUAN Wei-jiang. Gene Identification and Functional Analysis of Yellowish and Early Heading Mutant hz1 in Rice[J]. Biotechnology Bulletin, 2024, 40(7): 125-136.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 内切酶 Endonuclease |
|---|---|---|
| SE5-GFP-F | GTTGGTACCCGCTATAAGAGGGAGAGAGG | Kpn Ⅰ |
| SE5-GFP-R | CCGTCTAGAGGTGAATATGTGACGGAGGA | Xba Ⅰ |
| LB-R | ACGATGGACTCCAGTCCGGCCcttgaccaactctatcagagcttgg | |
| HZ1-F | CAAGGACCAGGCCAAGGAAG | |
| HZ1-R | ACCAAACTCAAAACAGGGGG | |
| InDel1-F | CAGGTATTCTGAGGTTGTATCC | |
| InDel1-R | CACGGTCAAATTCAACATTCC | |
| dCAP3-F | CACCAACTGAGCTCACTAGCCATAT | NdeⅠ |
| dCAP3-R | GGCCCAATGACTACCTTCTACTTTA | |
| SE5-ex-F | GGCTGAAAAGGACTCCCAAG | |
| SE5-ex-R | GTCCAGCTAGAGGCGACTTC | |
| Actin1-ex-F | GACTCTGGTGATGGTGTCAGC | |
| Actin1-ex-R | GGCTGGAAGAGGACCTCAGG | |
| Hd3a-ex-F | TTGGTAGGGTTGTGGGTGATGTGC | |
| Hd3a-ex-R | AGGTTAGGGTCACTTGGGCTTGGT | |
| RFT1-ex-F | TCCGAGCCCAAGCAACCCTAAC | |
| RFT1-ex-R | AGTTCCTGGTGCTGAAGTTCTG | |
| HEMA-ex-F | GATGCAATCACTGCTGGAAAGCGT | |
| HEMA-ex-R | CCATCTTGCCAGCACCAATCAACA | |
| PORA-ex-F | TCGTCGGCCTCGTCTGAGTTTATT | |
| PORA-ex-R | AGGCCTCTCTCACTGAAAGCTGAA | |
| YGL1-ex-F | CCAGCCACTGATGAAAGCAGCAAT | |
| YGL1-ex-R | AGAGCGCTAATACACTCGCGAACA | |
| CAO1-ex-F | CTTGTCGTATTCTTGGCGAG | |
| CAO1-ex-R | ATCCCGTGATGCTGTCGCTA |
表1 引物序列
Table 1 Primer sequence
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 内切酶 Endonuclease |
|---|---|---|
| SE5-GFP-F | GTTGGTACCCGCTATAAGAGGGAGAGAGG | Kpn Ⅰ |
| SE5-GFP-R | CCGTCTAGAGGTGAATATGTGACGGAGGA | Xba Ⅰ |
| LB-R | ACGATGGACTCCAGTCCGGCCcttgaccaactctatcagagcttgg | |
| HZ1-F | CAAGGACCAGGCCAAGGAAG | |
| HZ1-R | ACCAAACTCAAAACAGGGGG | |
| InDel1-F | CAGGTATTCTGAGGTTGTATCC | |
| InDel1-R | CACGGTCAAATTCAACATTCC | |
| dCAP3-F | CACCAACTGAGCTCACTAGCCATAT | NdeⅠ |
| dCAP3-R | GGCCCAATGACTACCTTCTACTTTA | |
| SE5-ex-F | GGCTGAAAAGGACTCCCAAG | |
| SE5-ex-R | GTCCAGCTAGAGGCGACTTC | |
| Actin1-ex-F | GACTCTGGTGATGGTGTCAGC | |
| Actin1-ex-R | GGCTGGAAGAGGACCTCAGG | |
| Hd3a-ex-F | TTGGTAGGGTTGTGGGTGATGTGC | |
| Hd3a-ex-R | AGGTTAGGGTCACTTGGGCTTGGT | |
| RFT1-ex-F | TCCGAGCCCAAGCAACCCTAAC | |
| RFT1-ex-R | AGTTCCTGGTGCTGAAGTTCTG | |
| HEMA-ex-F | GATGCAATCACTGCTGGAAAGCGT | |
| HEMA-ex-R | CCATCTTGCCAGCACCAATCAACA | |
| PORA-ex-F | TCGTCGGCCTCGTCTGAGTTTATT | |
| PORA-ex-R | AGGCCTCTCTCACTGAAAGCTGAA | |
| YGL1-ex-F | CCAGCCACTGATGAAAGCAGCAAT | |
| YGL1-ex-R | AGAGCGCTAATACACTCGCGAACA | |
| CAO1-ex-F | CTTGTCGTATTCTTGGCGAG | |
| CAO1-ex-R | ATCCCGTGATGCTGTCGCTA |
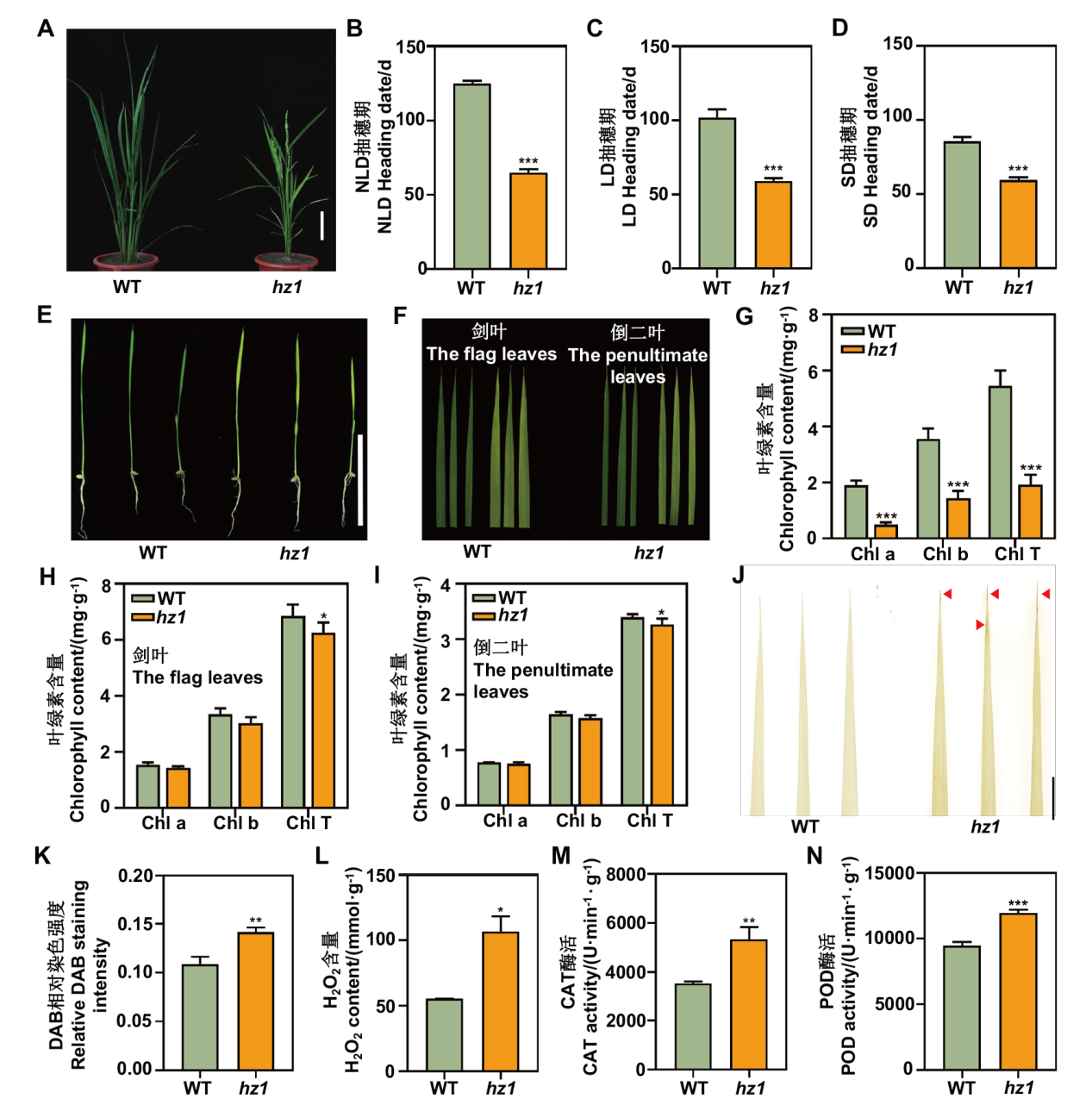
图1 hz1突变体的表型分析 A:hz1突变体在田间种植条件下的表型,比例尺为10 cm;B-D:分别为田间自然长日照(NLD)、长日照(LD)、短日照(SD)条件下WT与hz1的抽穗期,数值表示为平均值±SD(n≥25);E:苗期hz1的表型,比例尺为1 cm;F:抽穗期hz1叶片表型;G:苗期hz1的叶绿素含量测定,数值表示为平均值±SD(n=3);H:抽穗期hz1的剑叶叶绿素含量测定,数值表示为平均值±SD(n=3);I:抽穗期hz1的倒二叶叶绿素含量测定,数值表示为平均值±SD(n=3)(Chl a:叶绿素a;Chl b:叶绿素b;Chl T:总叶绿素含量);J:hz1的DAB染色结果,红色箭头所示红褐色斑点,比例尺为1 cm;K:hz1的DAB染色相对强弱分析,数值表示为平均值±SD(n=3);L:hz1突变体叶片过氧化氢含量的测定,数值表示为平均值±SD(n=3);M和N:hz1突变体叶片过氧化氢酶CAT及过氧化物酶POD的酶活性测定,数值表示为平均值±SD(n=3)。*,**,***分别表示在0.05、0.01和0.001水平上显著性差异,下同
Fig. 1 Phenotype analysis of hz1 mutant A: The phenotype of hz1 mutant in the field, bar=10 cm. B-D: The heading dates of hz1 under natural long day(NLD), long day(LD), and short day(SD)conditions. Values are shown as mean ± SD(n ≥ 25). E: The phenotype of hz1 during the seedling stage, bar=1 cm. F: The leaves of hz1 at heading stage. G: Measurement of chlorophyll content during seedling stage. Values are shown as mean ± SD(n=3). H: The chlorophyll content of flag leaves during the heading stage. Values are shown as mean ± SD(n=3). I: The chlorophyll content of the penultimate leaves. Values are shown as mean ± SD(n=3)(Chl a: chlorophyll a; Chl b: chlorophyll b; Chl T: total chlorophyll content). J: The DAB staining results of WT and hz1. Red arrows indicate brown spots, bar=1 cm. K: Relative intensity of DAB staining for WT and hz1. Values are shown as mean ± SD(n=3). L: The measurement of hydrogen peroxide(H2O2)content in leaves. Values are shown as mean ± SD(n=3). M and N: Enzyme activity measurements of catalase(CAT)and peroxidase(POD)in leaves. Values are shown as mean ± SD(n=3). *: P<0.05; **: P<0.01; ***: P<0.001, the same below

图2 hz1突变体的农艺性状分析 A-G:分别为株高、分蘖数、穗长、一次枝梗数、二次枝梗数、结实率及百粒重的统计分析,数值表示为平均值±SD(n=30);H:hz1的穗型,比例尺为10 cm;I:hz1的籽粒数及结实率,比例尺为10 cm;J和 K:hz1的花粉染色分析,放大倍数为400倍;箭头所示为典型败育花粉
Fig. 2 Analysis of agronomic traits of hz1 mutant A-G: Statistical analysis of plant height, tiller number, panicle length, number of primary branches, number of secondary branches, seed setting rate, and 100-grain weight. Values are shown as mean ± SD(n=30). H: The panicle of hz1, bar=10 cm. I: Grain number and seed setting rate of hz1, bar=10 cm. J and K: The pollen staining of hz1. The magnified times are 400 times, and the arrows indicate abortive pollens
| 组合 Cross | 正常表型植株数 Number of wild plants | 突变表型植株数 Number of mutant plants | 总植株数 Number of total plants | ꭕ2(3∶1) | P value |
|---|---|---|---|---|---|
| ZH11/hz1 | 458 | 130 | 588 | 2.621 3 | 0.105 4 |
表2 突变体hz1的遗传分析
Table 2 Genetic analysis of hz1 Mutant
| 组合 Cross | 正常表型植株数 Number of wild plants | 突变表型植株数 Number of mutant plants | 总植株数 Number of total plants | ꭕ2(3∶1) | P value |
|---|---|---|---|---|---|
| ZH11/hz1 | 458 | 130 | 588 | 2.621 3 | 0.105 4 |
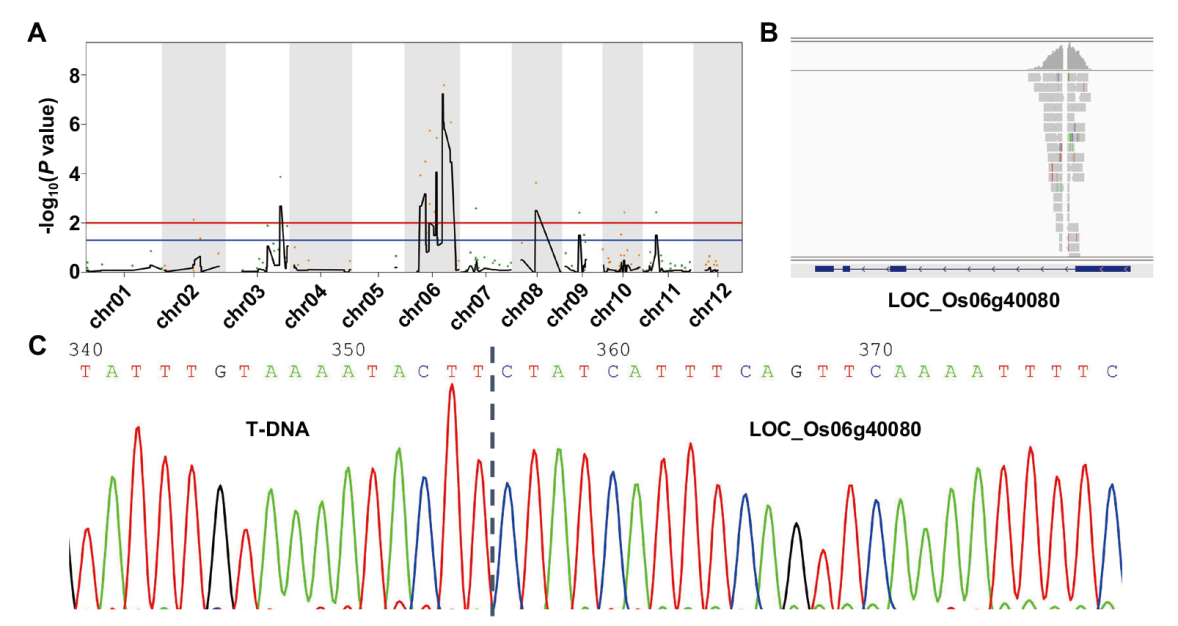
图3 BSA-seq分析及基因定位 A:BSA-seq分析,HZ1与水稻第6染色体上的分子标记紧密连锁;B:T-DNA插入位点与HZ1共分离;C:T-DNA插入位点测序结果
Fig. 3 BSA-seq analysis and gene mapping A: BSA-seq analysis. HZ1 is closely linked with the molecular marker on rice chromosome 6. B: T-DNA insertion site is co-segregated with HZ1. C: Sequencing analysis of T-DNA insertion site
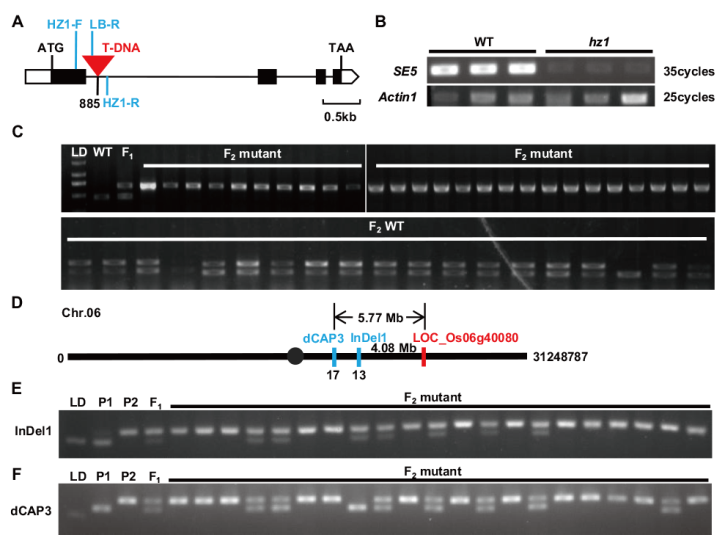
图4 T-DNA插入位点的检测及连锁分析 A:LOC_Os06g40080位点及T-DNA插入的位置示意图,HZ1-F、HZ1-R及LB-R所示为检测引物位置;B:HZ1/SE5基因RT-PCR表达分析,Actin1为水稻内参基因;C:T-DNA插入位点的检测及连锁分析,LD为DL 2000 marker,WT为野生型,F1为F1植株,F2 mutant和F2 WT分别为F2代群体中突变表型植株及野生型植株;D:水稻第6染色体及InDel1、dCAP3分子标记位置示意图;E和F:InDel1及dCAP3标记PCR扩增结果,LD为DL 2000 marker,P1为亲本ZH11植株,P2为亲本hz1植株,F1为F1植株,F2 mutant为F2代群体中突变表型植株
Fig. 4 Detection and linkage analysis of T-DNA insertion sites A: Schematic diagram of LOC_Os06g40080 site and T-DNA insertion location. HZ1-F, HZ1-R, and LB-R indicate the positions of the detection primers. B: RT-PCR analysis of HZ1/SE5 gene. Actin1 is an internal control in rice. C: The linkage analysis of T-DNA insertion sites. LD is DL 2000 marker, WT is wild-type, F1 is F1 plant, F2 mutant and F2 WT are plants with mutant phenotype and WT phenotype in the F2 population, respectively. D: Schematic diagram of InDel1, dCAP3 markers in the chromosome 6. E and F: PCR amplification of InDel1 and dCAP3. LD is DL 2000 marker, P1 is ZH11 plant, P2 is hz1 mutant, F1 is the F1 plants, and F2 mutant are the plants with hz1 phenotype in the F2 population
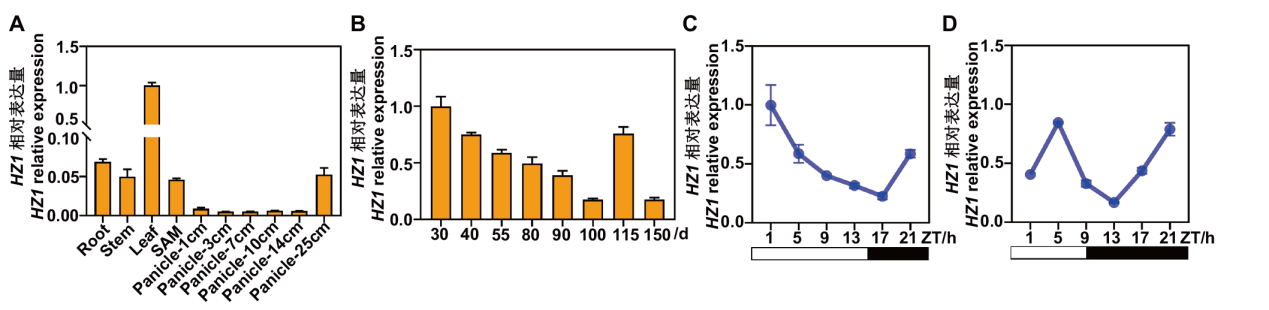
图5 HZ1/SE5基因表达模式分析 A:HZ1/SE5组织特异性表达分析(SAM:茎顶端分生组织);B:HZ1/SE5在水稻不同生长时期的表达分析;C和D分别为HZ1/SE5在LD(C)及SD(D)条件下的表达分析,ZT(zeitgeber time, h)表示给光时间。白色空心方框表示光期,黑色实心方框表示暗期。下同
Fig. 5 Expression patterns of HZ1/SE5 gene A: Tissue-specific expression analysis of HZ1/SE5(SAM: stem apical meristem). B: Expression analysis of HZ1/SE5 at different growth stages in rice. C and D: Expression analysis of HZ1/SE5 under LD (C) and SD (D) conditions, respectively. ZT(zeitgeber time, h)indicates the time of light on. The white hollow box indicates the light period, and the black solid box indicates the dark period. The same below
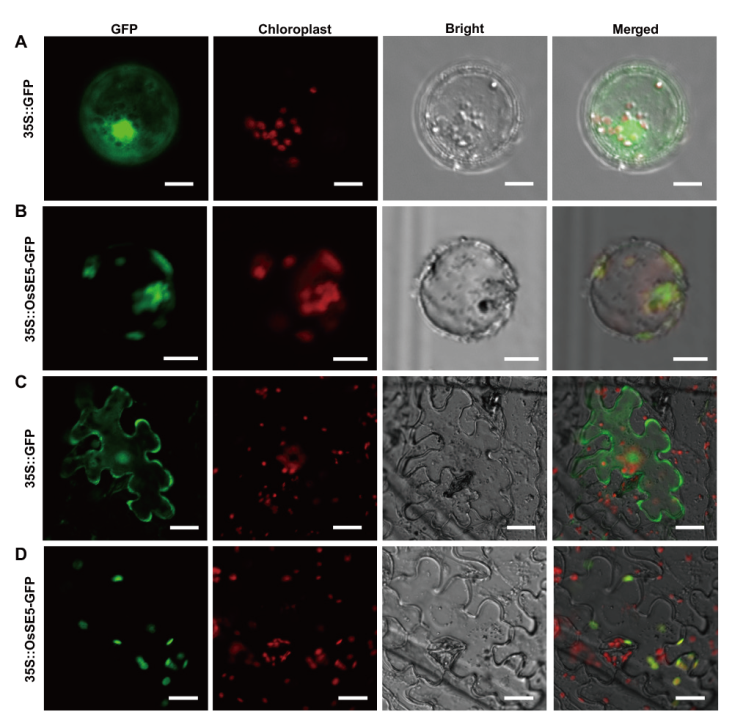
图6 HZ1/SE5蛋白的亚细胞定位 A和B:空载体和HZ1-GFP融合蛋白在水稻原生质体中的瞬时表达,比例尺为10 μm;C和D:空载体和HZ1-GFP融合蛋白在烟草表皮细胞中的瞬时表达,比例尺为20 μm
Fig. 6 Subcellular localization of HZ1/SE5 protein A and B: Transient expression of empty vector and HZ1-GFP fusion protein in rice protoplasts, bar=10 μm. C and D: Transient expression of empty vector and HZ1-GFP fusion protein in tobacco epidermal cells, bar=20 μm
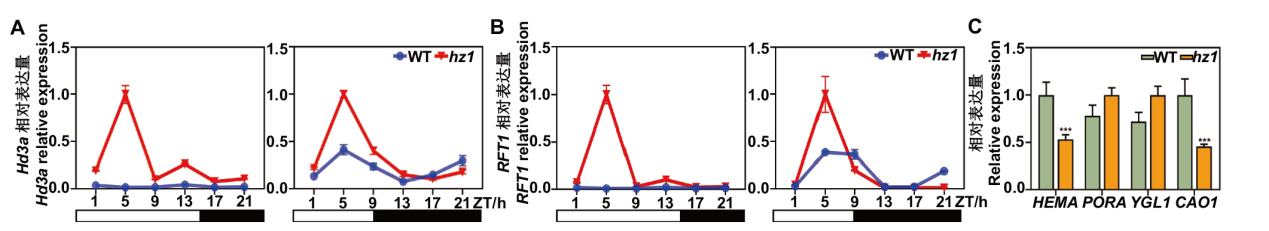
图7 光周期调控通路关键基因与叶绿素合成相关基因在hz1突变体中的表达分析 A, B:LD及SD条件下,光周期调控通路中关键基因在WT及hz1突变体中的表达水平;C:叶绿素合成相关基因在WT与hz1突变体中的表达
Fig. 7 Expression analysis of key genes involved in photoperiodic regulatory pathway and chlorophyll synthesis related genes in hz1 mutant A, B: Expression of key genes associated with photoperiodic regulatory pathway in WT and hz1 mutants under LD and SD conditions. C: Expression of chlorophyll synthesis related genes in WT and hz1 mutants
| [1] |
Yano M, Katayose Y, Ashikari M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS[J]. Plant Cell, 2000, 12(12): 2473-2484.
doi: 10.1105/tpc.12.12.2473 pmid: 11148291 |
| [2] | Lin HX, Yamamoto T, Sasaki T, et al. Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines[J]. Theor Appl Genet, 2000, 101(7): 1021-1028. |
| [3] |
Kojima S, Takahashi Y, Kobayashi Y, et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions[J]. Plant Cell Physiol, 2002, 43(10): 1096-1105.
doi: 10.1093/pcp/pcf156 pmid: 12407188 |
| [4] | Zhu YJ, Fan YY, Wang K, et al. Rice Flowering Locus T 1 plays an important role in heading date influencing yield traits in rice[J]. Sci Rep, 2017, 7(1): 4918. |
| [5] |
Komiya R, Ikegami A, Tamaki S, et al. Hd3a and RFT1 are essential for flowering in rice[J]. Development, 2008, 135(4): 767-774.
doi: 10.1242/dev.008631 pmid: 18223202 |
| [6] |
Xue WY, Xing YZ, Weng XY, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice[J]. Nat Genet, 2008, 40(6): 761-767.
doi: 10.1038/ng.143 pmid: 18454147 |
| [7] | Casal JJ, Luccioni LG, Oliverio KA, et al. Light, phytochrome signalling and photomorphogenesis in Arabidopsis[J]. Photochem Photobiol Sci, 2003, 2(6): 625-636. |
| [8] |
Wang X, Jiang BC, Gu LF, et al. A photoregulatory mechanism of the circadian clock in Arabidopsis[J]. Nat Plants, 2021, 7(10): 1397-1408.
doi: 10.1038/s41477-021-01002-z pmid: 34650267 |
| [9] |
Kami C, Lorrain S, Hornitschek P, et al. Light-regulated plant growth and development[J]. Curr Top Dev Biol, 2010, 91: 29-66.
doi: 10.1016/S0070-2153(10)91002-8 pmid: 20705178 |
| [10] |
Kliebenstein DJ, Lim JE, Landry LG, et al. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1[J]. Plant Physiol, 2002, 130(1): 234-243.
doi: 10.1104/pp.005041 pmid: 12226503 |
| [11] | Cheng MC, Kathare PK, Paik I, et al. Phytochrome signaling networks[J]. Annu Rev Plant Biol, 2021, 72: 217-244. |
| [12] |
Shekhawat GS, Verma K. Haem oxygenase(HO): an overlooked enzyme of plant metabolism and defence[J]. J Exp Bot, 2010, 61(9): 2255-2270.
doi: 10.1093/jxb/erq074 pmid: 20378668 |
| [13] |
Davis SJ, Kurepa J, Vierstra RD. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases[J]. Proc Natl Acad Sci USA, 1999, 96(11): 6541-6546.
doi: 10.1073/pnas.96.11.6541 pmid: 10339624 |
| [14] |
Izawa T, Oikawa T, Tokutomi S, et al. Phytochromes confer the photoperiodic control of flowering in rice(a short-day plant)[J]. Plant J, 2000, 22(5): 391-399.
doi: 10.1046/j.1365-313x.2000.00753.x pmid: 10849355 |
| [15] |
Andrés F, Galbraith DW, Talón M, et al. Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice[J]. Plant Physiol, 2009, 151(2): 681-690.
doi: 10.1104/pp.109.139097 pmid: 19675157 |
| [16] |
Chen H, Cheng ZJ, Ma XD, et al. A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice[J]. Plant Cell Rep, 2013, 32(12): 1855-1867.
doi: 10.1007/s00299-013-1498-y pmid: 24043333 |
| [17] | Peng YL, Zou T, Li LM, et al. Map-based cloning and functional analysis of YE1 in rice, which is involved in light-dependent chlorophyll biogenesis and photoperiodic flowering pathway[J]. Int J Mol Sci, 2019, 20(3): 758. |
| [18] | Rao YC, Xu N, Li SF, et al. PE-1, encoding heme oxygenase 1, impacts heading date and chloroplast development in rice(Oryza sativa L.)[J]. J Agric Food Chem, 2019, 67(26): 7249-7257. |
| [19] |
Arnon DI. Copper enzymes in isolated chloroplasts. polyphenoloxidase in beta vulgaris[J]. Plant Physiol, 1949, 24(1): 1-15.
doi: 10.1104/pp.24.1.1 pmid: 16654194 |
| [20] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [21] |
Trinidad JL, Longkumer T, Kohli A. Rice protoplast isolation and transfection for transient gene expression analysis[J]. Methods Mol Biol, 2021, 2238: 313-324.
doi: 10.1007/978-1-0716-1068-8_21 pmid: 33471341 |
| [22] | Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis[J]. Plant Cell, 1991, 3(11): 1177-1186. |
| [23] |
Terry MJ, Kendrick RE. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato[J]. Plant Physiol, 1999, 119(1): 143-152.
pmid: 9880355 |
| [24] | Kraepiel Y, Jullien M, Cordonnier-Pratt MM, et al. Identification of two loci involved in phytochrome expression in Nicotiana plumbaginifolia and lethality of the corresponding double mutant[J]. Mol Gen Genet, 1994, 242(5): 559-565. |
| [25] | Weller JL, Terry MJ, Rameau C, et al. The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXα[J]. Plant Cell, 1996, 8(1): 55-67. |
| [26] |
Davis SJ, Bhoo SH, Durski AM, et al. The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants[J]. Plant Physiol, 2001, 126(2): 656-669.
doi: 10.1104/pp.126.2.656 pmid: 11402195 |
| [27] | Legris M, Ince YÇ, Fankhauser C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants[J]. Nat Commun, 2019, 10(1): 5219. |
| [28] |
Pham VN, Kathare PK, Huq E. Phytochromes and phytochrome interacting factors[J]. Plant Physiol, 2018, 176(2): 1025-1038.
doi: 10.1104/pp.17.01384 pmid: 29138351 |
| [29] |
Osugi A, Itoh H, Ikeda-Kawakatsu K, et al. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice[J]. Plant Physiol, 2011, 157(3): 1128-1137.
doi: 10.1104/pp.111.181792 pmid: 21880933 |
| [30] | Wu HM, Zheng Y, Liu J, et al. Heme oxygenase-1 delays gibberellin-induced programmed cell death of rice aleurone layers subjected to drought stress by interacting with nitric oxide[J]. Front Plant Sci, 2016, 6: 1267. |
| [31] |
Hsu YY, Chao YY, Kao CH. Biliverdin-promoted lateral root formation is mediated through heme oxygenase in rice[J]. Plant Signal Behav, 2012, 7(7): 885-887.
doi: 10.4161/psb.20458 pmid: 22751314 |
| [32] | Xu S, Wang LJ, Zhang B, et al. RNAi knockdown of rice SE5 gene is sensitive to the herbicide methyl viologen by the down-regulation of antioxidant defense[J]. Plant Mol Biol, 2012, 80(2): 219-235. |
| [33] | Xie YJ, Mao Y, Xu S, et al. Heme-heme oxygenase 1 system is involved in ammonium tolerance by regulating antioxidant defence in Oryza sativa[J]. Plant Cell Environ, 2015, 38(1): 129-143. |
| [34] | Meng FY, Feng NJ, Zheng DF, et al. Exogenous Hemin alleviates NaCl stress by promoting photosynthesis and carbon metabolism in rice seedlings[J]. Sci Rep, 2023, 13(1): 3497. |
| [1] | 沈真辉, 曹瑶, 杨林雷, 罗祥英, 子灵山, 陆青青, 李荣春. 金耳和毛韧革菌麦角硫因生物合成基因的克隆及生物信息学分析[J]. 生物技术通报, 2024, 40(7): 259-272. |
| [2] | 黄丹, 姜山, 彭涛. 褐角苔FfCYP98基因克隆及其功能分析[J]. 生物技术通报, 2024, 40(7): 273-284. |
| [3] | 田胜尼, 张琴, 董玉飞, 丁洲, 叶爱华, 张明珠. 酸性矿山废水对成熟期水稻根区理化因子及固氮微生物的影响[J]. 生物技术通报, 2024, 40(6): 271-280. |
| [4] | 王玉书, 赵琳琳, 赵爽, 胡琦, 白慧霞, 王欢, 曹业萍, 范震宇. 大白菜BrCYP83B1基因的克隆及表达分析[J]. 生物技术通报, 2024, 40(6): 152-160. |
| [5] | 郝思怡, 张君珂, 王斌, 曲朋燕, 李瑞得, 程春振. 香蕉ELF3的克隆与表达分析[J]. 生物技术通报, 2024, 40(5): 131-140. |
| [6] | 杜泽光, 任少文, 张凤勤, 李梅兰, 李改珍, 齐仙惠. 大白菜BrMLP328的克隆、表达及功能验证[J]. 生物技术通报, 2024, 40(4): 122-129. |
| [7] | 刘换换, 杨立春, 李火根. 北美鹅掌楸LtMYB305基因的克隆及功能分析[J]. 生物技术通报, 2024, 40(4): 179-188. |
| [8] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [9] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [10] | 李兴容, 谭志兵, 赵燕, 李曜魁, 赵炳然, 唐丽. 水稻低亲和性阳离子转运蛋白基因OsLCT3的克隆与功能研究[J]. 生物技术通报, 2024, 40(4): 97-109. |
| [11] | 李雪, 李容欧, 孔美懿, 黄磊. 解淀粉芽孢杆菌SQ-2对水稻的促生作用[J]. 生物技术通报, 2024, 40(2): 109-119. |
| [12] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [13] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [14] | 邹修为, 岳佳妮, 李志宇, 戴良英, 李魏. 水稻热激转录因子HsfA2b调控非生物胁迫抗性的功能分析[J]. 生物技术通报, 2024, 40(2): 90-98. |
| [15] | 张超, 王子瑞, 孙亚丽, 毛馨晨, 唐家琪, 于恒秀. 水稻维生素B1合成相关基因OsTHIC的功能研究[J]. 生物技术通报, 2024, 40(2): 99-108. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||