








生物技术通报 ›› 2025, Vol. 41 ›› Issue (2): 331-342.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0090
• 研究报告 • 上一篇
收稿日期:2024-01-23
出版日期:2025-02-26
发布日期:2025-02-28
通讯作者:
雷艳丽,女,实验师,研究方向 :中药活性物质基础与药效学评价;E-mail: 912567436@qq.com作者简介:黎伟华,女,实验师,研究方向 :医学实验动物学及病理学;E-mail: 546240233@qq.com
基金资助:
LI Wei-hua1( ), WU Jing1, JIN Xue-qin2, LEI Yan-li1(
), WU Jing1, JIN Xue-qin2, LEI Yan-li1( )
)
Received:2024-01-23
Published:2025-02-26
Online:2025-02-28
摘要:
目的 应用标记定量(TMT)蛋白质组学技术,探究化学诱导剂(carbon tetrachloride,CCl4)致小鼠急性肝损伤的作用机制。 方法 20只SPF级雄性BALB/C小鼠随机分为对照组和诱导组,每组10只,腹腔注射诱导8周(2次/周)。通过测定血液生化水平及肝组织病理切片等指标评价肝损伤的程度。通过串联质谱标记技术(TMT)进行蛋白质组学分析,对发现的差异蛋白进行基因本体论(GO)注释、KEGG通路和蛋白互作网络分析。 结果 与对照组比较,CCl4诱导组的血液生化指标谷草转氨酶(aspartate aminotransferase,AST)的值显著升高(P<0.05);谷丙转氨酶(alanine aminotransferase,ALT)、谷氨酰转肽酶(Y-glutamyl transpeptadase,GGT)、总胆汁酸(total biliary acid,TBA)的值极显著增加(P<0.01),且肝组织病理切片显示CCl4诱导组小鼠肝损伤明显。利用TMT技术共鉴定了5 955个蛋白,其中440个蛋白强度增加,294个蛋白强度降低,差异蛋白在蛋白表达差异倍数、生物标志物应用和参与经典信号通路(JAK-STAT、PI3K-Akt、Rap1、GnRH、AGE-RAGE)中发挥重要作用。经典信号通路中差异蛋白存在显著富集,且富集程度相对较高。 结论 CCl4作为肝损伤评价的理想化学诱导剂,参与了机体免疫、细胞分裂、凋亡和自噬、炎症和肿瘤形成等多个关键过程,多方向多靶点还原了肝损伤的病理机制,为保肝药物的研发提供了坚实的理论基础和视角。
黎伟华, 吴璟, 金学琴, 雷艳丽. 基于蛋白质组学方法探讨四氯化碳诱导的小鼠急性肝损伤的差异蛋白表达[J]. 生物技术通报, 2025, 41(2): 331-342.
LI Wei-hua, WU Jing, JIN Xue-qin, LEI Yan-li. Exploring the Relative Differential Protein Expression of Carbon Tetrachloride-induced Acute Liver Injury in Mice Based on the Proteomics Method[J]. Biotechnology Bulletin, 2025, 41(2): 331-342.

图1 比较组肝功能相关血清生化指标和肝脏超声切片A,B:比较组的肝实质回声;C:比较组右肝最大斜位直径的变化;D-H:比较组的ALT、AST、ALP、GGT和TBA的表达水平
Fig. 1 Liver function-related serum biochemical indexes and liver ultrasound sections in the comparison groupsA, B: The liver parenchyma echo in the comparison group. C: Changes in the maximum oblique position diameter of the right liver in the comparison group. D-H: The expressions of ALT, AST, ALP, GGT, and TBA in the comparison groups. *P<0.05, **P<0.01, ***P<0.001

图2 CCl4诱导引起大体形态和组织病理学改变A,B:比较组肝大体形态图;C:比较组肝指数统计结果(n=6);D,E:比较组肝组织H&E染色;F,G:比较组肝组织Masson染色
Fig. 2 CCl4 induction caused gross morphological and histopathological changes (200×)A, B: Plot of gross liver morphology in the comparison group. C: Liver index statistics in the comparison group (n=6). D-E: H & E staining of liver tissue in the comparison groups. F-G: The Masson staining of liver sections's in the comparison groups

图3 比较组差异蛋白鉴定结果和差异蛋白的GO注释图A:比较组差异显著性火山图;B:比较组差异蛋白在BP、CC、MF(-log10(P值)>8.0)生物过程分类下GO功能富集图;C:比较组前5个上调蛋白GO项图;D:比较组下调蛋白的前5个GO项图
Fig. 3 Results of differential protein identification and the GO annotation plots of the differential proteins in comparison groupA: Volcano map of the significant difference in the comparison group. B: GO functional enrichment plots of comparison group differential proteins under BP, CC, and MF (-log10 (P-value)>8.0) biological process classification. C: GO terms plots of the top five up regulated proteins in the comparison group. D: GO terms plots of the top five down regulated proteins in the comparison group
调节(上调/下调) Regulation(Up/Down) | 蛋白编号Accession | 功能描述 Description | 基因符号Gene symbol | 分子量Mw/kD | 比值(CCl4/Control) Ratio(CCl4/Control) | P值 P-value |
|---|---|---|---|---|---|---|
| Up | P50236 | Bile salt sulfotransferase 2 | Sult2a2 | 33.3 | 13.086 | 1.71E-04 |
| Q64449 | C-type mannose receptor 2 | Mrc2 | 167 | 5.077 | 3.11E-04 | |
| P13745 | Glutathione S-transferase A1 | Gsta1 | 25.6 | 4.575 | 2.36E-03 | |
| Q9R100 | Cadherin-17 | Cdh17 | 91.6 | 3.976 | 2.38E-04 | |
| P97501 | Dimethylaniline monooxygenase [N-oxide-forming] 3 | Fmo3 | 60.5 | 3.438 | 1.25E-03 | |
| Q6IS41 | Solute carrier family 25 member 47 | Slc25a47 | 33.6 | 3.224 | 2.49E-02 | |
| Q9CZS1 | Aldehyde dehydrogenase X, mitochondrial | Aldh1b1 | 57.5 | 3.108 | 1.66E-05 | |
| Q8BG95 | Protein phosphatase 1 regulatory subunit 12B | Ppp1r12b | 109 | 3.007 | 1.19E-02 | |
| Down | Q9QXS8 | Probable N-acetyltransferase CML5 | Cml5 | 25.7 | 0.089 | 2.98E-03 |
| Q61694 | NADPH-dependent 3-keto-steroid reductase Hsd3b5 | Hsd3b5 | 41.9 | 0.167 | 1.24E-03 | |
| Q63836 | Selenium-binding protein 2 | Selenbp2 | 52.6 | 0.156 | 4.23E-04 | |
| Q60991 | Cytochrome P450 7B1 | Cyp7b1 | 58.4 | 0.169 | 1.52E-04 | |
| Q9Z0N2 | Eukaryotic translation initiation factor 2 subunit 3, Y-linked | Eif2s3y | 51.1 | 0.21 | 6.87E-05 | |
| O35728 | Cytochrome P450 4A14 | Cyp4a14 | 58.7 | 0.221 | 1.30E-03 | |
| Q9QXZ6 | Solute carrier organic anion transporter family member 1A1 | Slco1a1 | 74.3 | 0.25 | 3.72E-04 | |
| Q61646 | Haptoglobin | Hp | 38.7 | 0.354 | 1.78E-02 | |
| P46425 | Glutathione S-transferase P 2 | Gstp2 | 23.5 | 0.393 | 1.37E-03 | |
| Q3UP75 | UDP-glucuronosyltransferase 3A1 | Ugt3a1 | 59.7 | 0.394 | 5.09E-04 |
表1 CCl4诱导后前10个表达上调和下调的蛋白
Table 1 The top ten up-and down-regulated differential proteins of CCl4-induced liver injury
调节(上调/下调) Regulation(Up/Down) | 蛋白编号Accession | 功能描述 Description | 基因符号Gene symbol | 分子量Mw/kD | 比值(CCl4/Control) Ratio(CCl4/Control) | P值 P-value |
|---|---|---|---|---|---|---|
| Up | P50236 | Bile salt sulfotransferase 2 | Sult2a2 | 33.3 | 13.086 | 1.71E-04 |
| Q64449 | C-type mannose receptor 2 | Mrc2 | 167 | 5.077 | 3.11E-04 | |
| P13745 | Glutathione S-transferase A1 | Gsta1 | 25.6 | 4.575 | 2.36E-03 | |
| Q9R100 | Cadherin-17 | Cdh17 | 91.6 | 3.976 | 2.38E-04 | |
| P97501 | Dimethylaniline monooxygenase [N-oxide-forming] 3 | Fmo3 | 60.5 | 3.438 | 1.25E-03 | |
| Q6IS41 | Solute carrier family 25 member 47 | Slc25a47 | 33.6 | 3.224 | 2.49E-02 | |
| Q9CZS1 | Aldehyde dehydrogenase X, mitochondrial | Aldh1b1 | 57.5 | 3.108 | 1.66E-05 | |
| Q8BG95 | Protein phosphatase 1 regulatory subunit 12B | Ppp1r12b | 109 | 3.007 | 1.19E-02 | |
| Down | Q9QXS8 | Probable N-acetyltransferase CML5 | Cml5 | 25.7 | 0.089 | 2.98E-03 |
| Q61694 | NADPH-dependent 3-keto-steroid reductase Hsd3b5 | Hsd3b5 | 41.9 | 0.167 | 1.24E-03 | |
| Q63836 | Selenium-binding protein 2 | Selenbp2 | 52.6 | 0.156 | 4.23E-04 | |
| Q60991 | Cytochrome P450 7B1 | Cyp7b1 | 58.4 | 0.169 | 1.52E-04 | |
| Q9Z0N2 | Eukaryotic translation initiation factor 2 subunit 3, Y-linked | Eif2s3y | 51.1 | 0.21 | 6.87E-05 | |
| O35728 | Cytochrome P450 4A14 | Cyp4a14 | 58.7 | 0.221 | 1.30E-03 | |
| Q9QXZ6 | Solute carrier organic anion transporter family member 1A1 | Slco1a1 | 74.3 | 0.25 | 3.72E-04 | |
| Q61646 | Haptoglobin | Hp | 38.7 | 0.354 | 1.78E-02 | |
| P46425 | Glutathione S-transferase P 2 | Gstp2 | 23.5 | 0.393 | 1.37E-03 | |
| Q3UP75 | UDP-glucuronosyltransferase 3A1 | Ugt3a1 | 59.7 | 0.394 | 5.09E-04 |
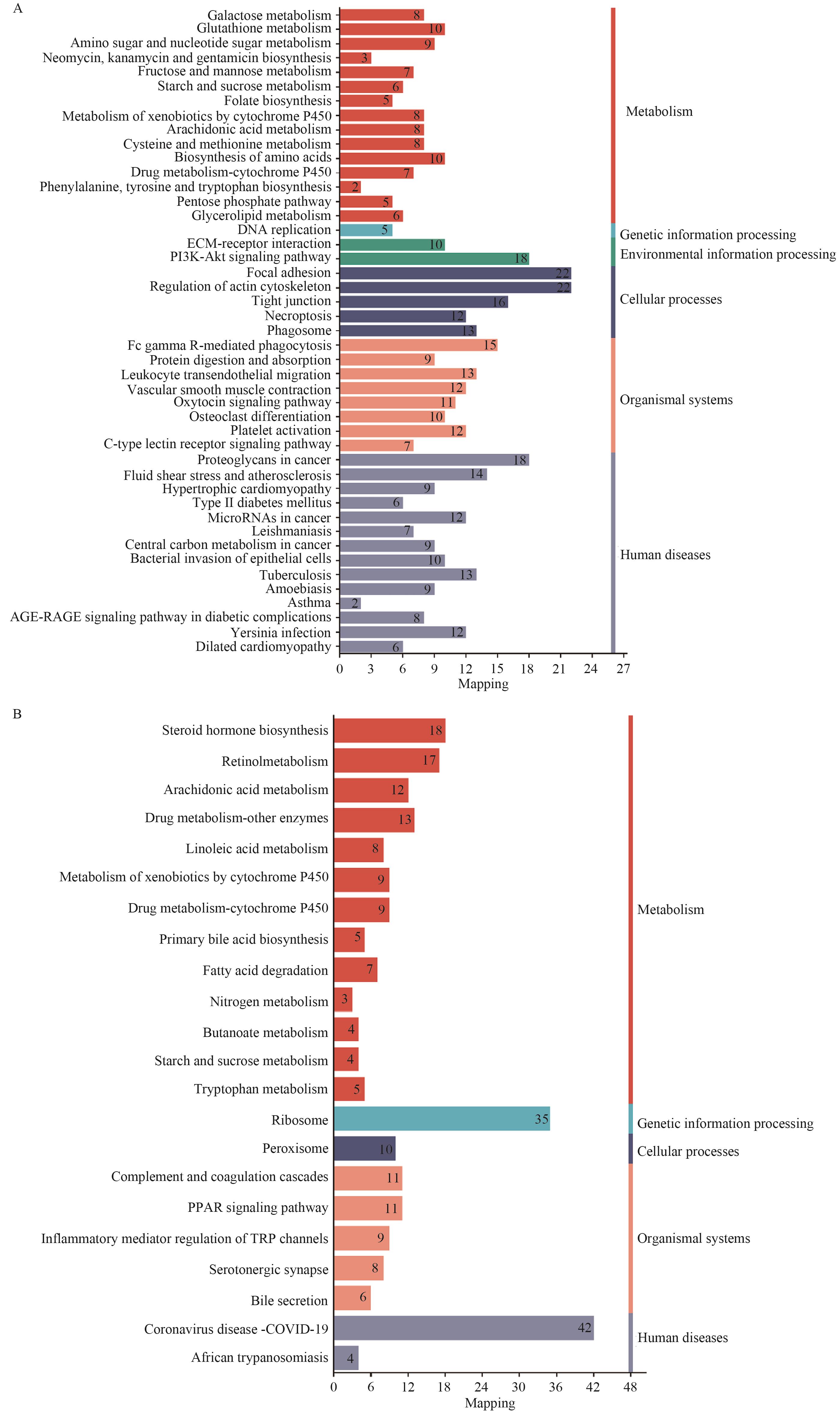
图4 上调和下调通路的KEGG通路分类和功能富集图A:上调通路;B:下调通路
Fig. 4 KEGG pathway classification and functional enrichment plots for up-regulated and down-regulated pathwaysA: Up-regulated pathway. B: Down-regulated pathway
疾病 Diseases | 相关的蛋白 Related proteins | 调节(上调/下调) Regulation(Up/Down) | 费希尔精确检验P值Fisher's exact test P value |
|---|---|---|---|
| Proteoglycans in cancer | P05480 Q63844 P51885 P20444 P49710 Q8BG95 P28654 P70336 P49817 P15379 Q01149 Q8BTM8 P70227 P26041 Q9JKF1 P26040 P11276 P11087 | Up | 7.58E-04 |
| Fluid shear stress and atherosclerosis | Q64337 P05480 P70236 P10648 P16460 Q09014 Q05144 Q64669 P61957 Q5EG47 P49817 Q99L20 P13745 Q60875 | Up | 1.45E-03 |
| Hypertrophic cardiomyopathy | Q6IRU2 P58771 Q60675 P82347 O54950 Q5EG47 A2ARA8 P31001 P09470 | Up | 1.53E-03 |
| MicroRNAs in cancer | P20152 P54227 Q63844 P58771 P63280 P20444 D3Z7P3 P21447 Q64261 P15379 P26645 P26040 | Up | 4.36E-03 |
| Tuberculosis | P05480 Q64449 Q63844 P04441 P19973 Q07813 P51437 O70370 P18242 P11835 O89053 P05555 P08508 | Up | 6.42E-03 |
| Leishmaniasis | Q63844 P28667 Q09014 P11835 Q61093 P05555 P08508 | Up | 5.11E-03 |
| Type II diabetes mellitus | Q63844 P52480 Q91W97 O08528 P28867 P17710 | Up | 2.66E-03 |
| C-type lectin receptor signaling pathway | Q9WVL2 P05480 Q63844 P19973 Q9WTK5 P28867 P70227 | Up | 3.56E-02 |
| Coronavirus disease-COVID-19 | P62918 O09167 P62852 P62849 P62267 P47915 P27659 P61514 P47964 P14115 P62754 P83882 Q9D8E6 P67984 P62830 P62281 Q9CPR4 Q6ZWV7 Q8BP67 P62301 P62751 P41105 P47963 Q9D823 Q9JJI8 P12970 P61255 P61358 Q6ZWV3 P47911 P84099 P62900 Q9D1R9 Q8K182 Q01279 Q8K0E8 Q8VCM7 Q8VCG4 E9PV24 P01029 Q8BH35 P62855 | Down | 1.72E-20 |
| African trypanosomiasis | Q00623 P02088 P02089 P01942 | Down | 1.11E-02 |
表2 KEGG分析中的相关疾病
Table 2 Related diseases in KEGG analysis
疾病 Diseases | 相关的蛋白 Related proteins | 调节(上调/下调) Regulation(Up/Down) | 费希尔精确检验P值Fisher's exact test P value |
|---|---|---|---|
| Proteoglycans in cancer | P05480 Q63844 P51885 P20444 P49710 Q8BG95 P28654 P70336 P49817 P15379 Q01149 Q8BTM8 P70227 P26041 Q9JKF1 P26040 P11276 P11087 | Up | 7.58E-04 |
| Fluid shear stress and atherosclerosis | Q64337 P05480 P70236 P10648 P16460 Q09014 Q05144 Q64669 P61957 Q5EG47 P49817 Q99L20 P13745 Q60875 | Up | 1.45E-03 |
| Hypertrophic cardiomyopathy | Q6IRU2 P58771 Q60675 P82347 O54950 Q5EG47 A2ARA8 P31001 P09470 | Up | 1.53E-03 |
| MicroRNAs in cancer | P20152 P54227 Q63844 P58771 P63280 P20444 D3Z7P3 P21447 Q64261 P15379 P26645 P26040 | Up | 4.36E-03 |
| Tuberculosis | P05480 Q64449 Q63844 P04441 P19973 Q07813 P51437 O70370 P18242 P11835 O89053 P05555 P08508 | Up | 6.42E-03 |
| Leishmaniasis | Q63844 P28667 Q09014 P11835 Q61093 P05555 P08508 | Up | 5.11E-03 |
| Type II diabetes mellitus | Q63844 P52480 Q91W97 O08528 P28867 P17710 | Up | 2.66E-03 |
| C-type lectin receptor signaling pathway | Q9WVL2 P05480 Q63844 P19973 Q9WTK5 P28867 P70227 | Up | 3.56E-02 |
| Coronavirus disease-COVID-19 | P62918 O09167 P62852 P62849 P62267 P47915 P27659 P61514 P47964 P14115 P62754 P83882 Q9D8E6 P67984 P62830 P62281 Q9CPR4 Q6ZWV7 Q8BP67 P62301 P62751 P41105 P47963 Q9D823 Q9JJI8 P12970 P61255 P61358 Q6ZWV3 P47911 P84099 P62900 Q9D1R9 Q8K182 Q01279 Q8K0E8 Q8VCM7 Q8VCG4 E9PV24 P01029 Q8BH35 P62855 | Down | 1.72E-20 |
| African trypanosomiasis | Q00623 P02088 P02089 P01942 | Down | 1.11E-02 |
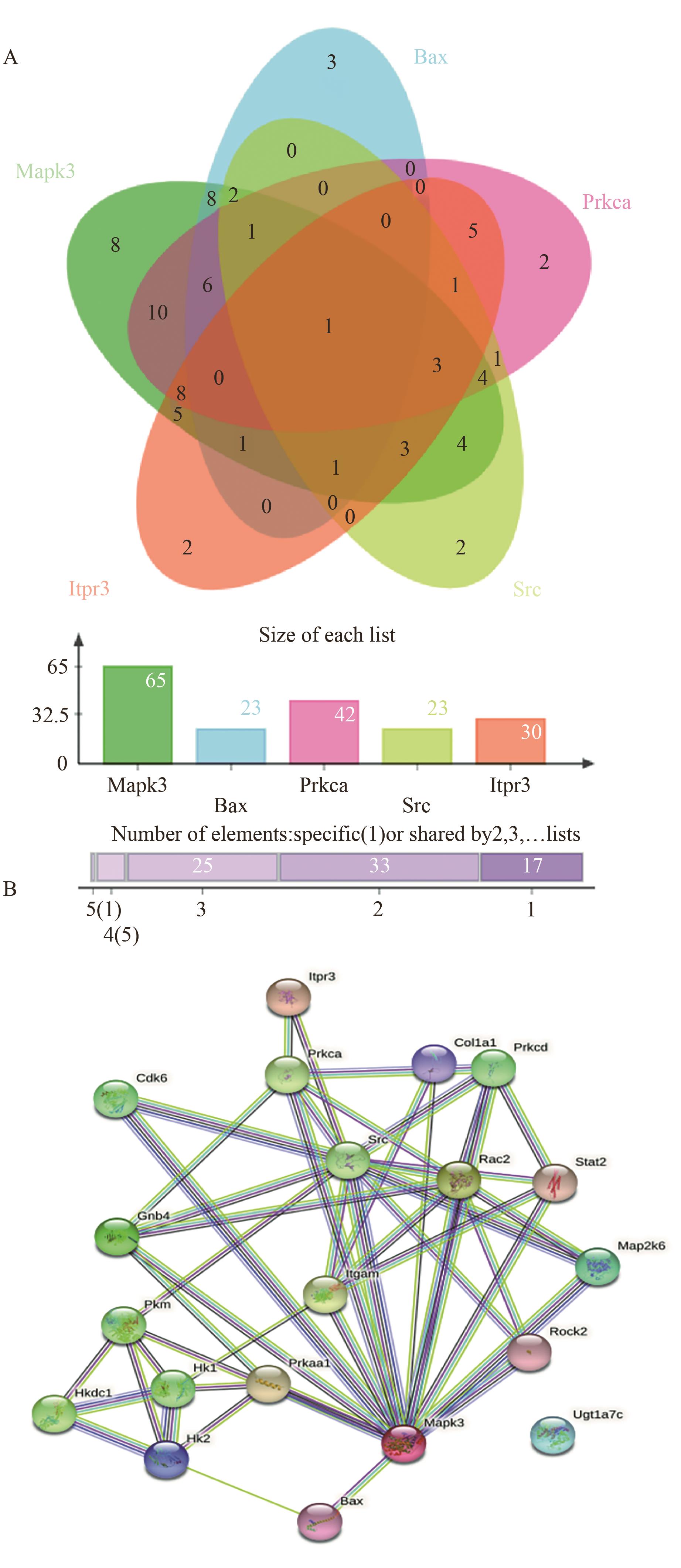
图5 蛋白质互作网络图A:差异显著蛋白连接程度文氏图;B:比较组DAPs网络互作图
Fig. 5 Protein interaction network diagramA: Degree of differential significant protein ligation Venn plot. B: DAPs network interaction map for comparison group
| 1 | Lachenmeier DW, Monakhova YB, Rehm J. Influence of unrecorded alcohol consumption on liver cirrhosis mortality [J]. World J Gastroenterol, 2014, 20(23): 7217-7222. |
| 2 | Okazaki I, Noro T, Tsutsui N, et al. Fibrogenesis and carcinogenesis in nonalcoholic steatohepatitis (NASH): involvement of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinase (TIMPs) [J]. Cancers, 2014, 6(3): 1220-1255. |
| 3 | Wang S, Lee Y, Kim J, et al. Potential role of Hedgehog pathway in liver response to radiation [J]. PLoS One, 2013, 8(9): e74141. |
| 4 | Xie GH, Karaca G, Swiderska-Syn M, et al. Cross-talk between Notch and Hedgehog regulates hepatic stellate cell fate in mice [J]. Hepatology, 2013, 58(5): 1801-1813. |
| 5 | Yilmaz Y, Eren F. Serum biomarkers of fibrosis and extracellular matrix remodeling in patients with nonalcoholic fatty liver disease: association with liver histology [J]. Eur J Gastroenterol Hepatol, 2019, 31(1): 43-46. |
| 6 | Luckey SW, Petersen DR. Activation of Kupffer cells during the course of carbon tetrachloride-induced liver injury and fibrosis in rats [J]. Exp Mol Pathol, 2001, 71(3): 226-240. |
| 7 | Gan DK, Zhang W, Huang CK, et al. Ursolic acid ameliorates CCl4-induced liver fibrosis through the NOXs/ROS pathway [J]. J Cell Physiol, 2018, 233(10): 6799-6813. |
| 8 | Xu GY, Han X, Yuan GX, et al. Screening for the protective effect target of deproteinized extract of calf blood and its mechanisms in mice with CCl4-induced acute liver injury [J]. PLoS One, 2017, 12(7): e0180899. |
| 9 | McCay PB, Lai EK, Poyer JL, et al. Oxygen- and carbon-centered free radical formation during carbon tetrachloride metabolism. Observation of lipid radicals in vivo and in vitro [J]. J Biol Chem, 1984, 259(4): 2135-2143. |
| 10 | Saijou E, Enomoto Y, Matsuda M, et al. Neutrophils alleviate fibrosis in the CCl4-induced mouse chronic liver injury model [J]. Hepatol Commun, 2018, 2(6): 703-717. |
| 11 | Li H, Zhang T, Wang K, et al. MFGE8 protects against CCl4-induced liver injury by reducing apoptosis and promoting proliferation of hepatocytes [J]. J Cell Physiol, 2019, 234(9): 16463-16474. |
| 12 | Zhao LD, Jin YH, Donahue K, et al. Tissue repair in the mouse liver following acute carbon tetrachloride depends on injury-induced Wnt/β-catenin signaling [J]. Hepatology, 2019, 69(6): 2623-2635. |
| 13 | Liu C, Tao Q, Sun MY, et al. Kupffer cells are associated with apoptosis, inflammation and fibrotic effects in hepatic fibrosis in rats [J]. Lab Invest, 2010, 90(12): 1805-1816. |
| 14 | D'Ambrosio R, Aghemo A, Rumi MG, et al. A morphometric and immunohistochemical study to assess the benefit of a sustained virological response in hepatitis C virus patients with cirrhosis [J]. Hepatology, 2012, 56(2): 532-543. |
| 15 | AlSaid M, Mothana R, Raish M, et al. Evaluation of the effectiveness of Piper cubeba extract in the amelioration of CCl4-induced liver injuries and oxidative damage in the rodent model [J]. Biomed Res Int, 2015, 2015: 359358. |
| 16 | Basu S. Carbon tetrachloride-induced lipid peroxidation: eicosanoid formation and their regulation by antioxidant nutrients [J]. Toxicology, 2003, 189(1-2): 113-127. |
| 17 | Park CM, Cha YS, Youn HJ, et al. Amelioration of oxidative stress by dandelion extract through CYP2E1 suppression against acute liver injury induced by carbon tetrachloride in Sprague-Dawley rats [J]. Phytother Res, 2010, 24(9): 1347-1353. |
| 18 | Son G, Iimuro Y, Seki E, et al. Selective inactivation of NF-kappaB in the liver using NF-kappaB decoy suppresses CCl4-induced liver injury and fibrosis [J]. Am J Physiol Gastrointest Liver Physiol, 2007, 293(3): G631-G639. |
| 19 | Tipoe GL, Leung TM, Liong EC, et al. Epigallocatechin-3-gallate (EGCG) reduces liver inflammation, oxidative stress and fibrosis in carbon tetrachloride (CCl4)-induced liver injury in mice [J]. Toxicology, 2010, 273(1-3): 45-52. |
| 20 | Hsieh CW, Ko WC, Ho WJ, et al. Antioxidant and hepatoprotective effects of Ajuga nipponensis extract by ultrasonic-assisted extraction [J]. Asian Pac J Trop Med, 2016, 9(5): 420-425. |
| 21 | Li SQ, Meng HY, Xi SM, et al. The effect of CCl4-induced acute liver injury on the ADAM8 expression in the mice [C]//2012 International Conference on Biomedical Engineering and Biotechnology. May 28-30, 2012: 589-592. |
| 22 | Vladimir-Knežević S, Cvijanović O, Blažeković B, et al. Hepatoprotective effects of Micromeria croatica ethanolic extract against CCl4-induced liver injury in mice [J]. BMC Complement Altern Med, 2015, 15: 233. |
| 23 | Ma JQ, Ding J, Zhang L, et al. Ursolic acid protects mouse liver against CCl4-induced oxidative stress and inflammation by the MAPK/NF-κB pathway [J]. Environ Toxicol Pharmacol, 2014, 37(3): 975-983. |
| 24 | Fu MZ, Yan YC, Su H, et al. Spleen proteome profiling of dairy goats infected with C. pseudotuberculosis by TMT-based quantitative proteomics approach [J]. J Proteomics, 2021, 248: 104352. |
| 25 | Klaas M, Kangur T, Viil J, et al. The alterations in the extracellular matrix composition guide the repair of damaged liver tissue [J]. Sci Rep, 2016, 6: 27398. |
| 26 | Hao YL, Fang HC, Zhao HL, et al. The role of microRNA-1 targeting of MAPK3 in myocardial ischemia-reperfusion injury in rats undergoing sevoflurane preconditioning via the PI3K/Akt pathway [J]. Am J Physiol Cell Physiol, 2018, 315(3): C380-388. |
| 27 | Runge-Morris M, Kocarek TA, Falany CN. Regulation of the cytosolic sulfotransferases by nuclear receptors [J]. Drug Metab Rev, 2013, 45(1): 15-33. |
| 28 | Mueller JW, Idkowiak J, Gesteira TF, et al. Human DHEA sulfation requires direct interaction between PAPS synthase 2 and DHEA sulfotransferase SULT2A1 [J]. J Biol Chem, 2018, 293(25): 9724-9735. |
| 29 | Bennett BJ, de Aguiar Vallim TQ, Wang ZN, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation [J]. Cell Metab, 2013, 17(1): 49-60. |
| 30 | Kovac S, Angelova PR, Holmström KM, et al. Nrf2 regulates ROS production by mitochondria and NADPH oxidase [J]. Biochim Biophys Acta, 2015, 1850(4): 794-801. |
| 31 | Xie YL, Chu JG, Jian XM, et al. Curcumin attenuates lipopolysaccharide/d-galactosamine-induced acute liver injury by activating Nrf2 nuclear translocation and inhibiting NF-kB activation [J]. Biomed Pharmacother, 2017, 91: 70-77. |
| 32 | Wang QQ, Zhang LL, Yuan XD, et al. The relationship between the bcl-2/bax proteins and the mitochondria-mediated apoptosis pathway in the differentiation of adipose-derived stromal cells into neurons [J]. PLoS One, 2016, 11(10): e0163327. |
| 33 | Yang XS, Li Q, Lin X, et al. Mechanism of fibrotic cardiomyopathy in mice expressing truncated Rho-associated coiled-coil protein kinase 1 [J]. FASEB J, 2012, 26(5): 2105-2116. |
| 34 | Yang L, Xiong ZY, Zhang LJ, et al. Tumor Necrosis Factor Alpha Down-regulated Human GSTA1 and GSTA4 Expression Through The NF-κB Signaling Pathway in Human Hepatoma HepG2 Cells [J]. Progress in Biochemistry and Biophysics, 2016, 43(8): 801-809. |
| 35 | Xie X, Peng J, Chang XT, et al. Activation of RhoA/ROCK regulates NF-κB signaling pathway in experimental diabetic nephropathy [J]. Mol Cell Endocrinol, 2013, 369(1-2): 86-97. |
| [1] | 李志强, 王吉英, 袁厅, 王佳, 韦艳娜, 王玉格, 李少丽, 邵国青, 冯志新, 于岩飞. 肺炎支原体感染评价方法的比较研究[J]. 生物技术通报, 2025, 41(1): 110-119. |
| [2] | 陈晓松, 刘超杰, 郑佳, 乔宗伟, 罗惠波, 邹伟. TMT定量蛋白质组学解析Rummeliibacillus suwonensis 3B-1 生长及己酸代谢机制[J]. 生物技术通报, 2024, 40(3): 135-145. |
| [3] | 高登科, 马白荣, 郭怡莹, 刘薇, 刘田, 靳亚平, 江舟, 陈华涛. 利用CRISPR/Cas9技术构建Quaking敲除的小鼠胚胎成纤维细胞株[J]. 生物技术通报, 2024, 40(2): 65-72. |
| [4] | 顾蕾, 张誉露, 唐尚睿, 于浩月, 李辰. 基于质谱的单细胞蛋白质组学技术的发展及应用[J]. 生物技术通报, 2024, 40(11): 125-141. |
| [5] | 许沛冬, 易剑锋, 陈迪, 陈浩, 谢丙炎, 赵文军. 组学技术在生防芽胞杆菌的应用进展[J]. 生物技术通报, 2024, 40(10): 208-220. |
| [6] | 丁亚群, 丁宁, 谢深民, 黄梦娜, 张昱, 张勤, 姜力. Vps28基因敲除小鼠模型的构建及其对泌乳和免疫性状影响的研究[J]. 生物技术通报, 2022, 38(3): 164-172. |
| [7] | 王智博, 王道平, 苗兰, 李瑛, 潘映红, 刘建勋. 血液样本蛋白质组分析方法的比较研究[J]. 生物技术通报, 2021, 37(8): 307-318. |
| [8] | 谢绍怡, 蒋璐蔓, 杨晓峰, 张仙玉, 吴珍芳, 李紫聪. 切割小鼠X染色体的CRISPR/Cas9系统表达载体的构建及验证[J]. 生物技术通报, 2021, 37(5): 67-75. |
| [9] | 张静, 熊燕, 华永琳, 郭玉, 熊显荣, 字向东, 李键. 小鼠骨骼肌纤维类型定量PCR内参基因的筛选[J]. 生物技术通报, 2021, 37(2): 71-79. |
| [10] | 孟丽娜, 彭春莹, 李铁栋, 李博生. 基于蛋白质组学对螺旋藻砷胁迫响应机制的研究[J]. 生物技术通报, 2020, 36(4): 107-116. |
| [11] | 李堃, 刘悦, 黄鹏, 杨智昉, 胡茜, 张颖, 李志宏, 吕叶辉, 梁乐. 小鼠精原细胞分化的蛋白质组学研究[J]. 生物技术通报, 2020, 36(3): 168-176. |
| [12] | 王棋文, 李盼, 潘翠云, 韩芬霞. 乙二醇对体内外源基因表达的作用研究[J]. 生物技术通报, 2019, 35(4): 64-68. |
| [13] | 张良, 陈小青, 宋佳宇, 毛然然, 姜倩雯, 林向民. 巴洛沙星胁迫下大肠杆菌的比较蛋白质组学研究[J]. 生物技术通报, 2019, 35(3): 103-109. |
| [14] | 张达秀, 贺双丽, 王倩, 蒲仕明, 吴琼. 青年和老年小鼠棕色脂肪来源间充质干细胞生物学特性比较[J]. 生物技术通报, 2019, 35(2): 137-142. |
| [15] | 耿慧君, 邹伟, 崔惠敬, 李晓宇, 王丽丽, 徐永平. 基于转录组学的金黄色葡萄球菌噬菌体安全性评估[J]. 生物技术通报, 2019, 35(12): 64-75. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||