








生物技术通报 ›› 2025, Vol. 41 ›› Issue (6): 87-98.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1215
吴浩1( ), 董伟峰1, 贺子天1, 李艳肖1, 谢辉2, 孙明哲1,3, 沈阳1,4(
), 董伟峰1, 贺子天1, 李艳肖1, 谢辉2, 孙明哲1,3, 沈阳1,4( ), 孙晓丽1(
), 孙晓丽1( )
)
收稿日期:2024-12-17
出版日期:2025-06-26
发布日期:2025-04-23
通讯作者:
沈阳,男,博士,研究方向 :寒地作物逆境生理与分子调控;E-mail: shenyang2024@byau.edu.cn作者简介:吴浩,男,硕士研究生,研究方向 :寒地作物逆境生理与分子调控;E-mail: 2864288928@qq.com
基金资助:
WU Hao1( ), DONG Wei-feng1, HE Zi-tian1, LI Yan-xiao1, XIE Hui2, SUN Ming-zhe1,3, SHEN Yang1,4(
), DONG Wei-feng1, HE Zi-tian1, LI Yan-xiao1, XIE Hui2, SUN Ming-zhe1,3, SHEN Yang1,4( ), SUN Xiao-li1(
), SUN Xiao-li1( )
)
Received:2024-12-17
Published:2025-06-26
Online:2025-04-23
摘要:
目的 BXL(β-D-xylosidase)属于β-木糖苷酶中第三族糖苷水解酶,负责催化细胞壁中木聚糖的降解,在植物生长发育及抵御非生物胁迫过程中发挥关键作用。鉴定和分析水稻OsBXL家族基因的进化及表达,为深入探究其功能奠定基础。 方法 通过生物信息学手段对OsBXL基因家族进行系统进化关系、复制事件、基因结构、保守基序及组织表达特征分析,并通过荧光定量PCR验证OsBXLs家族成员在非生物胁迫和激素处理下的表达变化情况。 结果 水稻BXL基因家族有10个成员,可划分为3个亚家族,其中Group III为单子叶植物特有;各亚族内的基因结构及保守结构域均表现出高度相似性;OsBXL基因分布在4条染色体上,其中4号染色体上的OsBXL5/6/7/8形成一个基因簇;转录组数据显示,OsBXL1/2/3/4/8在大多数组织中表现出较高的表达水平,而OsBXL5/6/7/10表达则相对较低;OsBXL7/10基因在特定发育阶段的幼穗和种子中高表达;OsBXL7在自然群体中存在3种不同单倍型,不同单倍型品种的籽粒长度、宽度和千粒重存在显著差异;荧光定量PCR结果显示,OsBXL家族成员对干旱、盐碱、脱落酸和茉莉酸甲酯处理的响应模式不同。 结论 水稻BXL基因家族具有高度的保守性,OsBXL7可能调控水稻籽粒大小,OsBXL1/3同时响应盐碱胁迫和茉莉酸甲酯处理。这些结果将为未来研究OsBXL基因功能提供重要参考。
吴浩, 董伟峰, 贺子天, 李艳肖, 谢辉, 孙明哲, 沈阳, 孙晓丽. 水稻BXL基因家族的全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(6): 87-98.
WU Hao, DONG Wei-feng, HE Zi-tian, LI Yan-xiao, XIE Hui, SUN Ming-zhe, SHEN Yang, SUN Xiao-li. Genome-wide Identification and Expression Analysis of the Rice BXL Gene Family[J]. Biotechnology Bulletin, 2025, 41(6): 87-98.
| 基因名称 Gene name | 上游引物 Forward primer (5'-3') | 下游引物 Reverse primer (5'-3') |
|---|---|---|
| OsBXL1 | GCTGACCAACCTCTACCTCAC | TGCCACCGAATTGACCTTCT |
| OsBXL2 | CACGTACAACCCTCCGTTCA | CCTCCAATCTCCCCTTGCAG |
| OsBXL3 | TACGGAGAGGAAGGCAGGAT | CTTGCACGACCTTCCTGGAT |
| OsBXL4 | AGCACTACACCGCATACGAC | AGCACATAACACTGGCCACA |
| OsBXL8 | AACGGTGTCCAACGCTACAA | GGCGCACATTATGCACGTAG |
| OsELf1-α | CATGATCACCGGTACCTCG | CCAGCATGTTGTCTCCGTG |
表1 荧光定量PCR引物信息
Table 1 Primers for quantitative fluorescence PCR
| 基因名称 Gene name | 上游引物 Forward primer (5'-3') | 下游引物 Reverse primer (5'-3') |
|---|---|---|
| OsBXL1 | GCTGACCAACCTCTACCTCAC | TGCCACCGAATTGACCTTCT |
| OsBXL2 | CACGTACAACCCTCCGTTCA | CCTCCAATCTCCCCTTGCAG |
| OsBXL3 | TACGGAGAGGAAGGCAGGAT | CTTGCACGACCTTCCTGGAT |
| OsBXL4 | AGCACTACACCGCATACGAC | AGCACATAACACTGGCCACA |
| OsBXL8 | AACGGTGTCCAACGCTACAA | GGCGCACATTATGCACGTAG |
| OsELf1-α | CATGATCACCGGTACCTCG | CCAGCATGTTGTCTCCGTG |

图1 OsBXLs蛋白保守结构域及序列比对A:蛋白结构域示意图;B:蛋白序列比对。SP:信号肽,Glyco_hydro_3 domain:糖苷水解酶N端催化结构域,Glyco_hydro_3_C domain:糖苷水解酶C端催化结构域,Fn 3 like domain:纤维连接蛋白Ⅲ型结构域;红色方框标注WGR和KH保守位点,蓝色三角形为亲核催化氨基酸残基位点Asp,黑色三角形为酸碱催化氨基酸残基位点Glu
Fig. 1 Functional domains and sequence alignment of OsBXL proteinsA: Schematic diagram of structural domains in proteins. B: Multiple sequence alignment of proteins; SP: signal peptide, Glyco_hydro_3 domain: N-terminal catalytic domain of glycoside hydrolase; Glyco_hydro_3_C domain: C-terminal catalytic domain of glycoside hydrolase; Fn 3-like domain: fibronectin type Ⅲ domain. red boxes mark the WGR and KH conserved sites, the blue triangle refers to the nucleophilic catalytic amino acid residue site Asp, the black triangle refers to the acid-base catalytic amino acid residue Glu
基因名称 Gene name | 基因登录号 Gene ID | 蛋白长度 Length of protein sequences (aa) | 分子量 Molecular weight (kD) | 理论等电点 Theoretical pI | 亲疏水性 GRAVY | 亚细胞定位/置信度 Subcellular location/Probability | 信号肽位置 Signal peptide position (aa) |
|---|---|---|---|---|---|---|---|
| OsBXL1 | LOC_Os01g19220 | 771 | 82.60 | 6.61 | -0.191 | 细胞壁/0.7852 Cell wall | 1-24 |
| OsBXL2 | LOC_Os02g51620 | 818 | 87.12 | 5.21 | 0.030 | 细胞壁/0.7618 Cell wall | 1-27 |
| OsBXL3 | LOC_Os04g44840 | 782 | 83.43 | 5.77 | -0.110 | 细胞壁/0.753 Cell wall | 1-20 |
| OsBXL4 | LOC_Os04g54810 | 770 | 83.65 | 5.84 | -0.111 | 细胞壁/0.8309 Cell wall | 1-26 |
| OsBXL5 | LOC_Os11g18690 | 793 | 85.03 | 6.33 | -0.103 | 细胞壁/0.5974 Cell wall | 1-26 |
| OsBXL6 | LOC_Os11g18730 | 780 | 84.30 | 6.32 | -0.146 | 细胞壁/0.8285 Cell wall | 1-22 |
| OsBXL7 | LOC_Os11g19160 | 853 | 91.66 | 9.10 | -0.130 | 细胞壁/0.7545 Cell wall | 1-21 |
| OsBXL8 | LOC_Os11g19210 | 816 | 88.75 | 6.19 | -0.150 | 细胞壁/0.6741 Cell wall | 1-31 |
| OsBXL9 | LOC_Os11g44950 | 765 | 80.92 | 6.42 | 0.022 | 细胞壁/0.8602 Cell wall | 1-18 |
| OsBXL10 | LOC_Os11g47350 | 664 | 70.19 | 7.92 | -0.069 | 细胞壁/0.7416 Cell wall | 1-24 |
表2 OsBXLs的基本信息
Table 2 Basic information of OsBXLs
基因名称 Gene name | 基因登录号 Gene ID | 蛋白长度 Length of protein sequences (aa) | 分子量 Molecular weight (kD) | 理论等电点 Theoretical pI | 亲疏水性 GRAVY | 亚细胞定位/置信度 Subcellular location/Probability | 信号肽位置 Signal peptide position (aa) |
|---|---|---|---|---|---|---|---|
| OsBXL1 | LOC_Os01g19220 | 771 | 82.60 | 6.61 | -0.191 | 细胞壁/0.7852 Cell wall | 1-24 |
| OsBXL2 | LOC_Os02g51620 | 818 | 87.12 | 5.21 | 0.030 | 细胞壁/0.7618 Cell wall | 1-27 |
| OsBXL3 | LOC_Os04g44840 | 782 | 83.43 | 5.77 | -0.110 | 细胞壁/0.753 Cell wall | 1-20 |
| OsBXL4 | LOC_Os04g54810 | 770 | 83.65 | 5.84 | -0.111 | 细胞壁/0.8309 Cell wall | 1-26 |
| OsBXL5 | LOC_Os11g18690 | 793 | 85.03 | 6.33 | -0.103 | 细胞壁/0.5974 Cell wall | 1-26 |
| OsBXL6 | LOC_Os11g18730 | 780 | 84.30 | 6.32 | -0.146 | 细胞壁/0.8285 Cell wall | 1-22 |
| OsBXL7 | LOC_Os11g19160 | 853 | 91.66 | 9.10 | -0.130 | 细胞壁/0.7545 Cell wall | 1-21 |
| OsBXL8 | LOC_Os11g19210 | 816 | 88.75 | 6.19 | -0.150 | 细胞壁/0.6741 Cell wall | 1-31 |
| OsBXL9 | LOC_Os11g44950 | 765 | 80.92 | 6.42 | 0.022 | 细胞壁/0.8602 Cell wall | 1-18 |
| OsBXL10 | LOC_Os11g47350 | 664 | 70.19 | 7.92 | -0.069 | 细胞壁/0.7416 Cell wall | 1-24 |

图2 水稻BXL家族系统进化关系分析绿色区域为Group Ⅰ、浅绿色区域为Group Ⅱ、橙色区域为Group Ⅲ
Fig. 2 Phylogenetic relation analysis of rice BXL familyThe green area indicates Group I, the light green area indicates Group II, and the orange area indicates Group Ⅲ

图3 OsBXLs基因结构和保守基序分析A:OsBXLs基因家族进化树;B:内含子-外显子结构组成,黄色方块代表CDS区、绿色方块代表UTR区域,实线代表内含子区域;C:保守基序,不同的motif对应关系位于图底部
Fig. 3 Gene structures and conserved motifs of OsBXLsA: Phylogenetic tree of the OsBXL genefamily. B: The intron-exon architecture, yellow boxes indicate CDS, green boxes indicate UTR, and solid lines indicate intron. C: Conserved motifs, different colored boxes at the bottom indicate different motifs
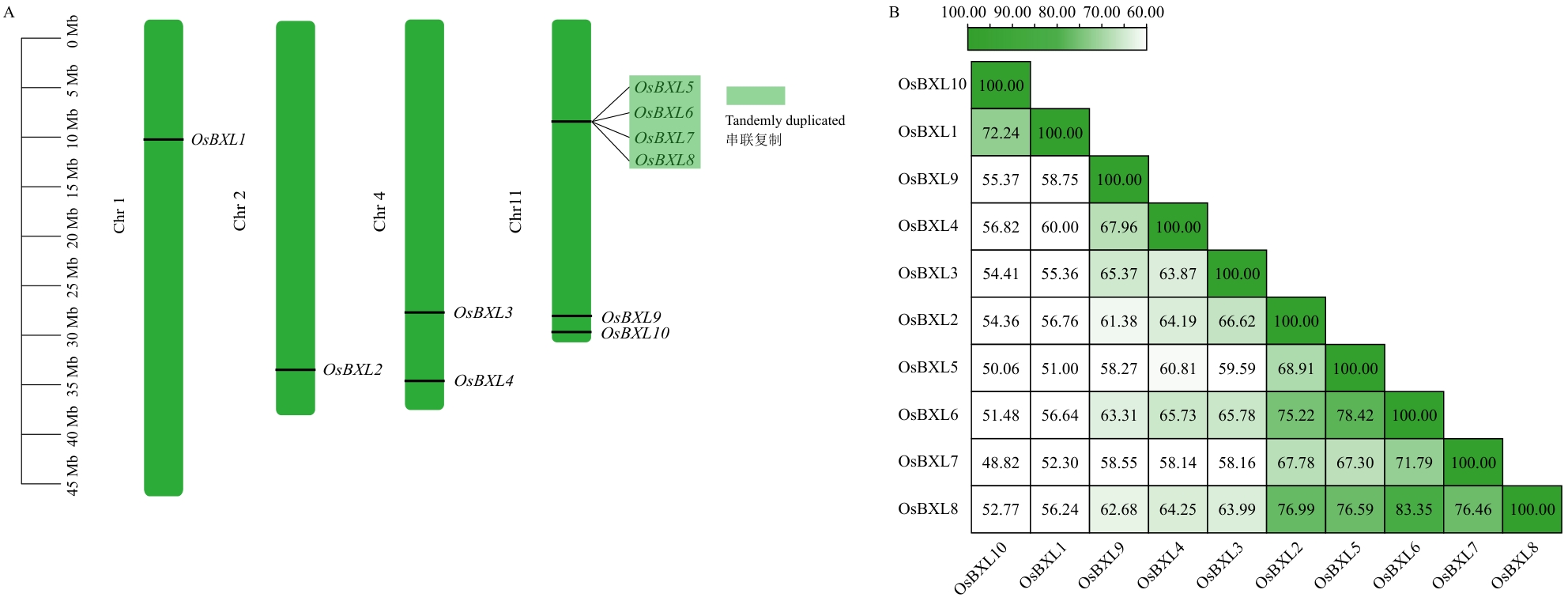
图4 OsBXLs基因染色体分布和蛋白序列相似度A:染色体定位,浅绿色区域显示4个OsBXLs基因簇;B:OsBXLs蛋白序列相似度,图中展示的数字表示氨基酸相似度,单位为百分比(%)
Fig. 4 Chromosomal location and protein sequence similarity of OsBXLsA: Chromosomal location, with the light green areas highlighting gene cluster containing four members. B: Protein sequence similarity of OsBXLs, with the numbers shown in the figure indicating amino acid similarity expressed as a percentage (%)
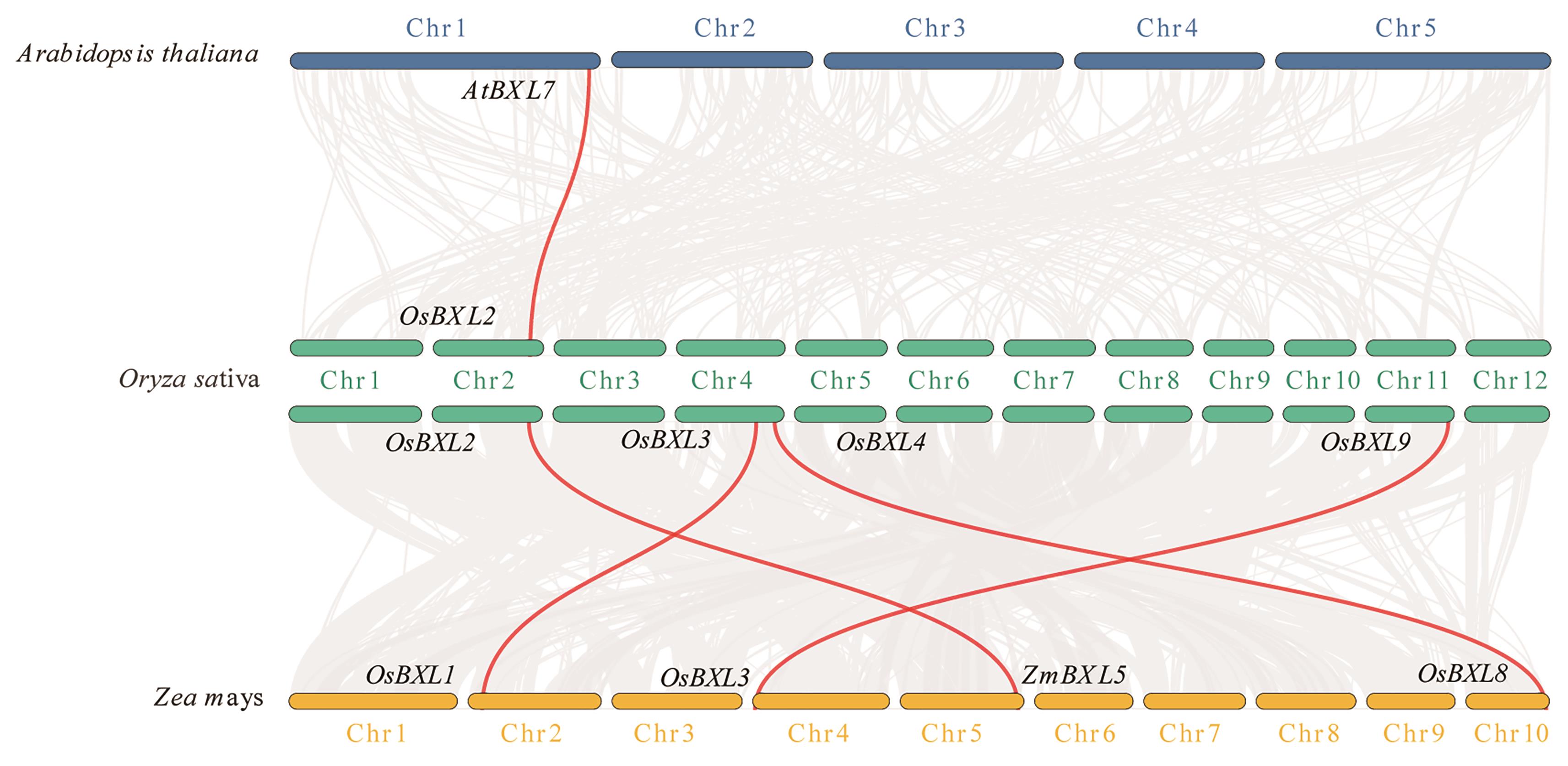
图5 水稻、玉米和拟南芥BXL家族基因共线性分析背景中灰线代表水稻与玉米和拟南芥基因组中的共线性基因对,红线代表OsBXLs与ZmBXLs和AtBXLs具有共线性的基因对;蓝色、绿色和黄色线条分别表示拟南芥、水稻和玉米的染色体
Fig. 5 Co-linearity analysis of BXL family genes in rice, maize, and ArabidopsisThe background gray lines indicate synteny in the genomes of rice with maize and Arabidopsis. The red lines indicate the synteny of OsBXLs with ZmBXLs and AtBXLs. The blue, green, and yellow bars indicate the chromosomes of Arabidopsis, rice, and maize, respectively

图6 OsBXLs家族成员的组织表达模式Seedling root:幼苗根;Mature leaf:成熟叶;Young leaf:幼叶;SAM:叶芽顶端分生组织;Inflorescence花序(P1:花序原基阶段;P2-P4:枝梗分化阶段;P5-P6:小穗分化阶段);Seed种子(S1:授粉后初期;S2-S3:胚发育阶段;S4-S5:籽粒成熟阶段)。红线区域对应Cluster I;绿线区域对应Cluster Ⅱ;将Log2FPKM >7定义为高表达;Log2FPKM <7定义为低表达
Fig. 6 Tissue expression pattern of OsBXLs genes family membersThe red line area corresponds to Cluster I, while the green line area corresponds to Cluster II; Log2FPKM >7 is defined as high expression, and Log2FPKM <7 is defined as low expression

图7 OsBXL7基因CDS区单倍型分析A:OsBXL7不同单倍型碱基及氨基酸变化示意图,CDS:编码序列,AA:编码氨基酸,REF:参考单倍型;B:不同OsBXL7单倍型的品种分布图;GJ:粳稻,XI:籼稻,admix:混合类型,Bas:基础品种;C-F:分别为不同OsBXL7单倍型的水稻籽粒长度、宽度、长宽比和千粒重;P值采用单因素方差分析计算
Fig. 7 Haplotype analysis of OsBXL7 based on the coding sequenceA: Schematic of the base and amino acid variations in different haplotypes of OsBXL7, CDS: coding sequence; AA: amino acids; REF: reference haplotype. B: Distribution of varieties with different haplotypes of OsBXL7. C-F: The grain length, grain width, length/width ratio and thousand-grain weight of rice varieties with different haplotypes of OsBXL7, respectively. P values were calculated by one-way ANOVA analysis
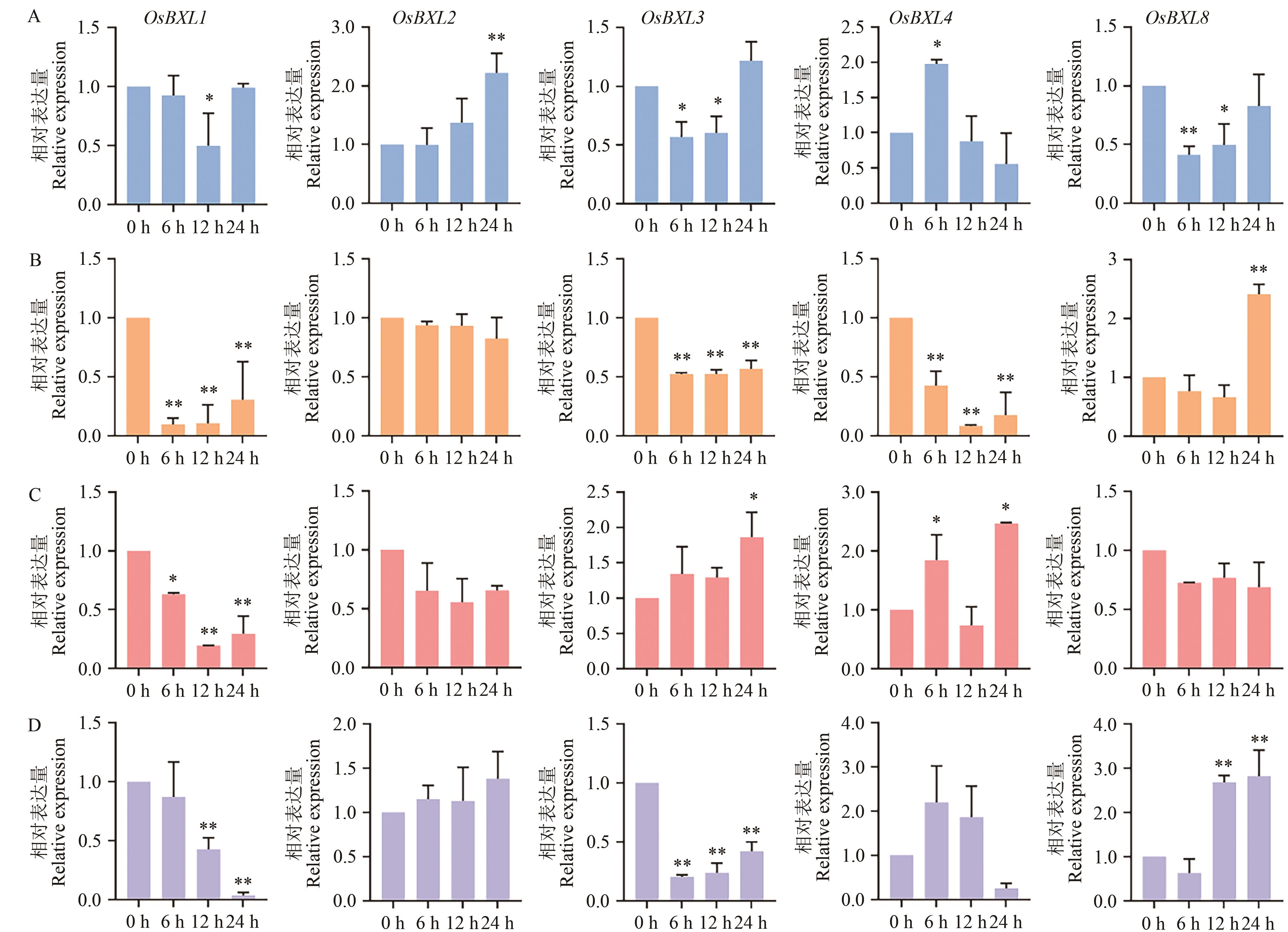
图8 OsBXLs基因在非生物胁迫和激素处理下的表达分析A:干旱胁迫;B:盐碱胁迫;C:ABA处理;D:MeJA处理;*,**分别表示P <0.05,P <0.01
Fig. 8 Expression analysis of OsBXLs under abiotic stress and hormone treatmentA: Drought stress. B: Salt-alkali stress. C: ABA treatment. D: MeJA treatment; * and ** indicate P <0.05, P <0.01, respectively
| [1] | Weidenhamer JD, Cipollini D, Morris K, et al. Ecological realism and rigor in the study of plant-plant allelopathic interactions [J]. Plant Soil, 2023, 489(1): 1-39. |
| [2] | Pandey P, Ramegowda V, Senthil-Kumar M. Shared and unique responses of plants to multiple individual stresses and stress combinations: physiological and molecular mechanisms [J]. Front Plant Sci, 2015, 6: 723. |
| [3] | Wan JX, He M, Hou QQ, et al. Cell wall associated immunity in plants [J]. Stress Biol, 2021, 1(1): 3. |
| [4] | 张曼, 张叶卓, 何其邹洪, 等. 植物细胞壁结构及成像技术研究进展 [J]. 生物技术通报, 2023, 39(7): 113-122. |
| Zhang M, Zhang YZ, He QZH, et al. Advances in plant cell wall structure and imaging technology [J]. Biotechnol Bull, 2023, 39(7): 113-122. | |
| [5] | Shtein I, Bar-On B, Popper ZA. Plant and algal structure: from cell walls to biomechanical function [J]. Physiol Plant, 2018, 164(1): 56-66. |
| [6] | Cleemput G, Hessing M, Van Oort M, et al. Purification and characterization of a β-D-xylosidase and an endo-xylanase from wheat flour [J]. Plant Physiol, 1997, 113(2): 377-386. |
| [7] | Li N, Zhang R, Zhou JP, et al. Structures, biochemical characteristics, and functions of β-xylosidases [J]. J Agric Food Chem, 2023, 71(21): 7961-7976. |
| [8] | Pontonio E, Mahony J, Di Cagno R, et al. Cloning, expression and characterization of a β-D-xylosidase from Lactobacillus rossiae DSM 15814(T) [J]. Microb Cell Fact, 2016, 15: 72. |
| [9] | Knob A, Terrasan CRF, Carmona EC. β-xylosidases from filamentous fungi: an overview [J]. World J Microbiol Biotechnol, 2010, 26(3): 389-407. |
| [10] | Minic Z, Rihouey C, Do CT, et al. Purification and characterization of enzymes exhibiting beta-D-xylosidase activities in stem tissues of Arabidopsis [J]. Plant Physiol, 2004, 135(2): 867-878. |
| [11] | Goujon T, Minic Z, El Amrani A, et al. AtBXL1, a novel higher plant (Arabidopsis thaliana) putative beta-xylosidase gene, is involved in secondary cell wall metabolism and plant development [J]. Plant J, 2003, 33(4): 677-690. |
| [12] | Chen JY, Qu CP, Chang RH, et al. Genome-wide identification of BXL genes in Populus trichocarpa and their expression under different nitrogen treatments [J]. 3 Biotech, 2020, 10(2): 57. |
| [13] | Lee RC, Hrmova M, Burton RA, et al. Bifunctional family 3 glycoside hydrolases from barley with α-l-arabinofuranosidase and β-d-xylosidase activity characterization, primary structures, and cooh-terminal processing [J]. J Biol Chem, 2003, 278(7): 5377-5387. |
| [14] | 赖彪, 陈春帆, 刘春渝, 等. 茎瘤芥BXL基因家族的全基因组鉴定及在瘤状茎膨大过程中的表达分析 [J]. 华中农业大学学报, 2020, 39(6): 155-163. |
| Lai B, Chen CF, Liu CY, et al. Genome-wide identification of BXL family genes in Brassica juncea var. tumida and their expression during swelling of stem [J]. J Huazhong Agric Univ, 2020, 39(6): 155-163. | |
| [15] | Carolina Di Santo M, Ilina N, Pagano EA, et al. A Japanese plum α-l-arabinofuranosidase/β-D-xylosidase gene is developmentally regulated by alternative splicing [J]. Plant Sci, 2015, 231: 173-183. |
| [16] | Collins T, Gerday C, Feller G. Xylanases, xylanase families and extremophilic xylanases [J]. FEMS Microbiol Rev, 2005, 29(1): 3-23. |
| [17] | Itai A, Yoshida K, Tanabe K, et al. A-D-xylosidase-like gene is expressed during fruit ripening in Japanese pear (Pyrus pyrifolia Nakai) [J]. J Exp Bot, 1999, 50(335): 877-878. |
| [18] | Bustamante CA, Civello PM, Martínez GA. Cloning of the promoter region of β-xylosidase (FaXyl1) gene and effect of plant growth regulators on the expression of FaXyl1 in strawberry fruit [J]. Plant Sci, 2009, 177(1): 49-56. |
| [19] | 丁文家, 胡峻铭, 王嘉力. 水稻育种主要目标性状基因挖掘研究进展 [J]. 杂交水稻, 2023, 38(3): 1-19. |
| Ding WJ, Hu JM, Wang JL. Research progress on gene mining of main target traits in rice breeding [J]. Hybrid Rice, 2023, 38(3): 1-19. | |
| [20] | 赵海成, 李红宇, 郑桂萍, 等. 寒地水稻新品种垦粳8号的选育及栽培技术 [J]. 黑龙江农业科学, 2021(1): 165-168. |
| Zhao HC, Li HY, Zheng GP, et al. Breeding and cultivation techniques of a new rice variety kenjing No.8 in cold region [J]. Heilongjiang Agric Sci, 2021(1): 165-168. | |
| [21] | Jain M, Nijhawan A, Arora R, et al. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress [J]. Plant Physiol, 2007, 143(4): 1467-1483. |
| [22] | Drula E, Garron ML, Dogan S, et al. The carbohydrate-active enzyme database: functions and literature [J]. Nucleic Acids Res, 2022, 50(D1): D571-D577. |
| [23] | Wang FM, Longkumer T, Catausan SC, et al. Genome-wide association and gene validation studies for early root vigour to improve direct seeding of rice [J]. Plant Cell Environ, 2018, 41(12): 2731-2743. |
| [24] | Guzha A, McGee R, Scholz P, et al. Cell wall-localized BETA-XYLOSIDASE4 contributes to immunity of Arabidopsis against Botrytis cinerea [J]. Plant Physiol, 2022, 189(3): 1794-1813. |
| [25] | 冯凯月, 赵鑫焱, 李子妍, 等. 植物响应盐碱胁迫的分子机制研究进展 [J]. 生物技术通报, 2024, 40(10): 122-138. |
| Feng KY, Zhao XY, Li ZY, et al. Research progress on molecular mechanism of plant response to saline-alkali stress [J]. Biotech.bull, 2024, 40(10): 122-138. | |
| [26] | 吴占清, 陈威, 赵展, 等. 玉米GRAS基因家族的全基因组鉴定及生物信息学分析 [J]. 中国农业科技导报, 2024, 26(3): 15-25. |
| Wu ZQ, Chen W, Zhao Z, et al. Genome-wide identification and bioinformatics analysis of GRAS gene family in maize [J]. J Agric Sci Technol, 2024, 26(3): 15-25. | |
| [27] | Cui D, Zhou H, Ma XD, et al. Genomic insights on the contribution of introgressions from Xian/Indica to the genetic improvement of Geng/Japonica rice cultivars [J]. Plant Commun, 2022, 3(3): 100325. |
| [28] | Ji C, Xu LN, Li YJ, et al. The O2-ZmGRAS11 transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize [J]. Mol Plant, 2022, 15(3): 468-487. |
| [29] | Bauer K, Nayem S, Lehmann M, et al. β-D-XYLOSIDASE 4 modulates systemic immune signaling in Arabidopsis thaliana [J]. Front Plant Sci, 2023, 13: 1096800. |
| [30] | Yu L, Wilson LFL, Terrett OM, et al. Evolution of glucuronoxylan side chain variability in vascular plants and the compensatory adaptations of cell wall-degrading hydrolases [J]. New Phytol, 2024, 244(3): 1024-1040. |
| [31] | Wang LQ, Guo K, Li Y, et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice [J]. BMC Plant Biol, 2010, 10: 282. |
| [32] | Yu XZ, Fan WJ, Lin YJ, et al. Differential expression of the PAL gene family in rice seedlings exposed to chromium by microarray analysis [J]. Ecotoxicology, 2018, 27(3): 325-335. |
| [33] | Sun LX, Deng RL, Liu JW, et al. An overview of sucrose transporter (SUT) genes family in rice [J]. Mol Biol Rep, 2022, 49(6): 5685-5695. |
| [34] | Xia D, Zhou H, Liu RJ, et al. GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice [J]. Mol Plant, 2018, 11(5): 754-756. |
| [35] | Liu JF, Chen J, Zheng XM, et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice [J]. Nat Plants, 2017, 3: 17043. |
| [36] | Gao XY, Zhang JQ, Zhang XJ, et al. Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling [J]. Plant Cell, 2019, 31(5): 1077-1093. |
| [37] | Zhu XY, Gou YJ, Heng YQ, et al. Targeted manipulation of grain shape genes effectively improves outcrossing rate and hybrid seed production in rice [J]. Plant Biotechnol J, 2023, 21(2): 381-390. |
| [38] | Nagashima Y, Ma ZY, Liu XT, et al. Multiple quality control mechanisms in the ER and TGN determine subcellular dynamics and salt-stress tolerance function of KORRIGAN1 [J]. Plant Cell, 2020, 32(2): 470-485. |
| [1] | 黄丹, 彭兵阳, 张盼盼, 焦悦, 吕佳斌. 油茶HD-Zip基因家族鉴定及其在非生物胁迫下的表达分析[J]. 生物技术通报, 2025, 41(6): 191-207. |
| [2] | 彭绍智, 王登科, 张祥, 戴雄泽, 徐昊, 邹学校. 辣椒CaFD1基因克隆、表达特征及功能验证[J]. 生物技术通报, 2025, 41(5): 153-164. |
| [3] | 刘源, 赵冉, 卢振芳, 李瑞丽. 植物类胡萝卜素生物代谢途径及其功能研究进展[J]. 生物技术通报, 2025, 41(5): 23-31. |
| [4] | 杜量衡, 唐黄磊, 张治国. 控制水稻光响应基因ELM1的图位克隆[J]. 生物技术通报, 2025, 41(5): 82-89. |
| [5] | 刘园园, 陈析丰, 钱前, 高振宇. 水稻穗发育调控的分子机制研究进展[J]. 生物技术通报, 2025, 41(5): 1-13. |
| [6] | 陈晓军, 惠建, 马洪文, 白海波, 钟楠, 李稼润, 樊云芳. 利用单碱基基因编辑技术创制OsALS抗除草剂水稻种质资源[J]. 生物技术通报, 2025, 41(4): 106-114. |
| [7] | 张益瑄, 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰. 马铃薯DIR家族全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(3): 123-136. |
| [8] | 韩江涛, 张帅博, 秦雅蕊, 韩硕洋, 张雅康, 王吉庆, 杜清洁, 肖怀娟, 李猛. 甜瓜β-淀粉酶基因家族的鉴定及对非生物胁迫的响应[J]. 生物技术通报, 2025, 41(3): 171-180. |
| [9] | 李欣芃, 张武汉, 张莉, 舒服, 何强, 郭杨, 邓华凤, 王悦, 孙平勇. γ射线诱变创制水稻突变体及其分子鉴定[J]. 生物技术通报, 2025, 41(3): 35-43. |
| [10] | 匡健华, 程志鹏, 赵永晶, 杨洁, 陈润乔, 陈龙清, 胡慧贞. 激素和非生物胁迫下荷花GH3基因家族的表达分析[J]. 生物技术通报, 2025, 41(2): 221-233. |
| [11] | 方慧敏, 顾艺枢, 张晶, 张龙. 水稻叶片淀粉的分离与理化性质分析[J]. 生物技术通报, 2025, 41(2): 51-57. |
| [12] | 葛仕杰, 刘怡德, 张华东, 宁强, 朱展望, 王书平, 刘易科. 小麦蛋白质二硫键异构酶基因家族的鉴定与表达[J]. 生物技术通报, 2025, 41(2): 85-96. |
| [13] | 殷缘, 程爽, 刘定豪, 邓晓霞, 李凯月, 王竞红, 蔺吉祥. 外源过氧化氢(H2O2)影响非生物胁迫下植物生长与生理代谢机制的研究进展[J]. 生物技术通报, 2025, 41(1): 1-13. |
| [14] | 武志健, 刘广洋, 林志豪, 盛彬, 陈鸽, 许晓敏, 王军伟, 徐东辉. 蔬菜种子萌发的纳米调控及其机制研究进展[J]. 生物技术通报, 2025, 41(1): 14-24. |
| [15] | 李禹欣, 李苗, 杜晓芬, 韩康妮, 连世超, 王军. 谷子SiSAP基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(1): 143-156. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||