








生物技术通报 ›› 2025, Vol. 41 ›› Issue (7): 150-163.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0018
李新妮1,2( ), 李俊怡1,2, 马雪华2, 何卫1,2, 李佳丽1,2, 于佳1,2, 曹晓宁1,2, 乔治军1,2(
), 李俊怡1,2, 马雪华2, 何卫1,2, 李佳丽1,2, 于佳1,2, 曹晓宁1,2, 乔治军1,2( ), 刘思辰1,2(
), 刘思辰1,2( )
)
收稿日期:2025-01-08
出版日期:2025-07-26
发布日期:2025-07-22
通讯作者:
:刘思辰,女,博士,副研究员,研究方向 :谷子种质资源评价利用;E-mail: lsch209@126.com作者简介:李新妮,女,硕士,研究方向 :作物遗传育种;E-mail: tangerine7423@163.com
基金资助:
LI Xin-ni1,2( ), LI Jun-yi1,2, MA Xue-hua2, HE Wei1,2, LI Jia-li1,2, YU Jia1,2, CAO Xiao-ning1,2, QIAO Zhi-jun1,2(
), LI Jun-yi1,2, MA Xue-hua2, HE Wei1,2, LI Jia-li1,2, YU Jia1,2, CAO Xiao-ning1,2, QIAO Zhi-jun1,2( ), LIU Si-chen1,2(
), LIU Si-chen1,2( )
)
Received:2025-01-08
Published:2025-07-26
Online:2025-07-22
摘要:
目的 PMEI(pectin methylesterase inhibitor)是控制细胞壁结构与特性的重要组成部分,在植物逆境胁迫响应方面发挥重要作用。探究谷子PMEI基因在非生物胁迫下的响应机制,为谷子的抗逆机理研究提供理论基础。 方法 运用生物信息学方法对谷子PMEI基因家族进行鉴定,采用实时荧光定量PCR技术对其家族成员在低温、干旱、MeJA、ABA胁迫下的表达模式进行分析。 结果 谷子基因组中共鉴定出68个SiPMEI基因家族成员,不均匀地分布在9条染色体上,大多数成员定位于细胞壁或叶绿体上。SiPMEI家族成员主要有2种结构域,PMEI结构域和pectinesteras+PMEI结构域,含有同一结构域的成员间理化性质、亚细胞定位和基因结构均较相似。启动子顺式作用元件分析表明SiPMEI基因家族成员含有多种非生物胁迫和激素响应元件。本研究选择了8个含有2种以上胁迫响应元件且含数量较多的SiPMEI家族成员进行实时荧光定量表达分析。结果表明,SiPMEI在谷子的根、茎、叶和穗中差异表达;非生物胁迫(低温、干旱)和激素胁迫(MeJA、ABA)处理下,SiPMEI基因的表达量在0‒24 h内整体呈现上升趋势,含PMEI结构域且定位于细胞壁上的成员SiPMEI30、SiPMEI32、SiPMEI36、SiPMEI63在胁迫下响应的最高表达量集中于8‒24 h。亚细胞定位于叶绿体上的成员SiPMEI22、SiPMEI31、SiPMEI38、SiPMEI47最高表达量分布较为分散,且上调位点较多。 结论 SiPMEI在低温、ABA和MeJA的胁迫下均具有正向响应,并在干旱和MeJA胁迫下具有类似的表达趋势。这些差异表达表明SiPMEI可能通过不同的分子机制响应非生物胁迫。
李新妮, 李俊怡, 马雪华, 何卫, 李佳丽, 于佳, 曹晓宁, 乔治军, 刘思辰. 谷子果胶甲酯酶抑制子PMEI基因家族鉴定及其对非生物胁迫的响应分析[J]. 生物技术通报, 2025, 41(7): 150-163.
LI Xin-ni, LI Jun-yi, MA Xue-hua, HE Wei, LI Jia-li, YU Jia, CAO Xiao-ning, QIAO Zhi-jun, LIU Si-chen. Identification of the PMEI Gene Family of Pectin Methylesterase Inhibitor in Foxtail Millet and Analysis of Its Response to Abiotic Stress[J]. Biotechnology Bulletin, 2025, 41(7): 150-163.
基因名称 Gene name | 基因序列 Gene sequence (5′‒3′) |
|---|---|
| 25S-F | AGGCAACAGAAACTCCATACG |
| 25S-R | ATGGCATAGCATTCATCACG |
| SiPMEI22-F | TGTTCTCGGGACAGGTTTCG |
| SiPMEI22-R | GTACAGCGTGTCCTGGTACG |
| SiPMEI30-F | GACCTCGGCTACGACTACTG |
| SiPMEI30-R | GGTTGTGCTCTTGGAGGCTA |
| SiPMEI31-F | TCTTCAGAGAGAGGGGGATCG |
| SiPMEI31-R | TAGCGCGACAAGCGAACAAT |
| SiPMEI32-F | TTTTCCGCGAACGTGTCCA |
| SiPMEI32-R | ACACCACCCCATATCGCTTG |
| SiPMEI36-F | CTCGGTACAGTCTTGCACCA |
| SiPMEI36-R | ATCCAGTGCAATGTCTCCGT |
| SiPMEI38-F | CCGAGTACATGAACCGTGGA |
| SiPMEI38-R | GTGGAGTTGAGCCAGAGGTC |
| SiPMEI47-F | CAGCCTTCGTGTTCCCTGTT |
| SiPMEI47-R | TCTTATTGATGACCGCCCTCG |
| SiPMEI63-F | AGCTTCGTTCGCTCATGCAA |
| SiPMEI63-R | AGTAATGAAGGCTGCAACACAC |
表1 用于RT-qPCR引物序列
Table 1 Primer sequences used for RT-qPCR
基因名称 Gene name | 基因序列 Gene sequence (5′‒3′) |
|---|---|
| 25S-F | AGGCAACAGAAACTCCATACG |
| 25S-R | ATGGCATAGCATTCATCACG |
| SiPMEI22-F | TGTTCTCGGGACAGGTTTCG |
| SiPMEI22-R | GTACAGCGTGTCCTGGTACG |
| SiPMEI30-F | GACCTCGGCTACGACTACTG |
| SiPMEI30-R | GGTTGTGCTCTTGGAGGCTA |
| SiPMEI31-F | TCTTCAGAGAGAGGGGGATCG |
| SiPMEI31-R | TAGCGCGACAAGCGAACAAT |
| SiPMEI32-F | TTTTCCGCGAACGTGTCCA |
| SiPMEI32-R | ACACCACCCCATATCGCTTG |
| SiPMEI36-F | CTCGGTACAGTCTTGCACCA |
| SiPMEI36-R | ATCCAGTGCAATGTCTCCGT |
| SiPMEI38-F | CCGAGTACATGAACCGTGGA |
| SiPMEI38-R | GTGGAGTTGAGCCAGAGGTC |
| SiPMEI47-F | CAGCCTTCGTGTTCCCTGTT |
| SiPMEI47-R | TCTTATTGATGACCGCCCTCG |
| SiPMEI63-F | AGCTTCGTTCGCTCATGCAA |
| SiPMEI63-R | AGTAATGAAGGCTGCAACACAC |

图1 SiPMEI序列特征分析A:SiPMEI基因家族成员进化树,B:保守基序分布;C:基因结构域分布;D:内含子与外显子结构
Fig. 1 Analysis of SiPMEI sequence characterizationA: SiPMEI evolutionary tree of gene family members; B: conserved motif distribution; C: gene domain distribution; D: intron and exon structure
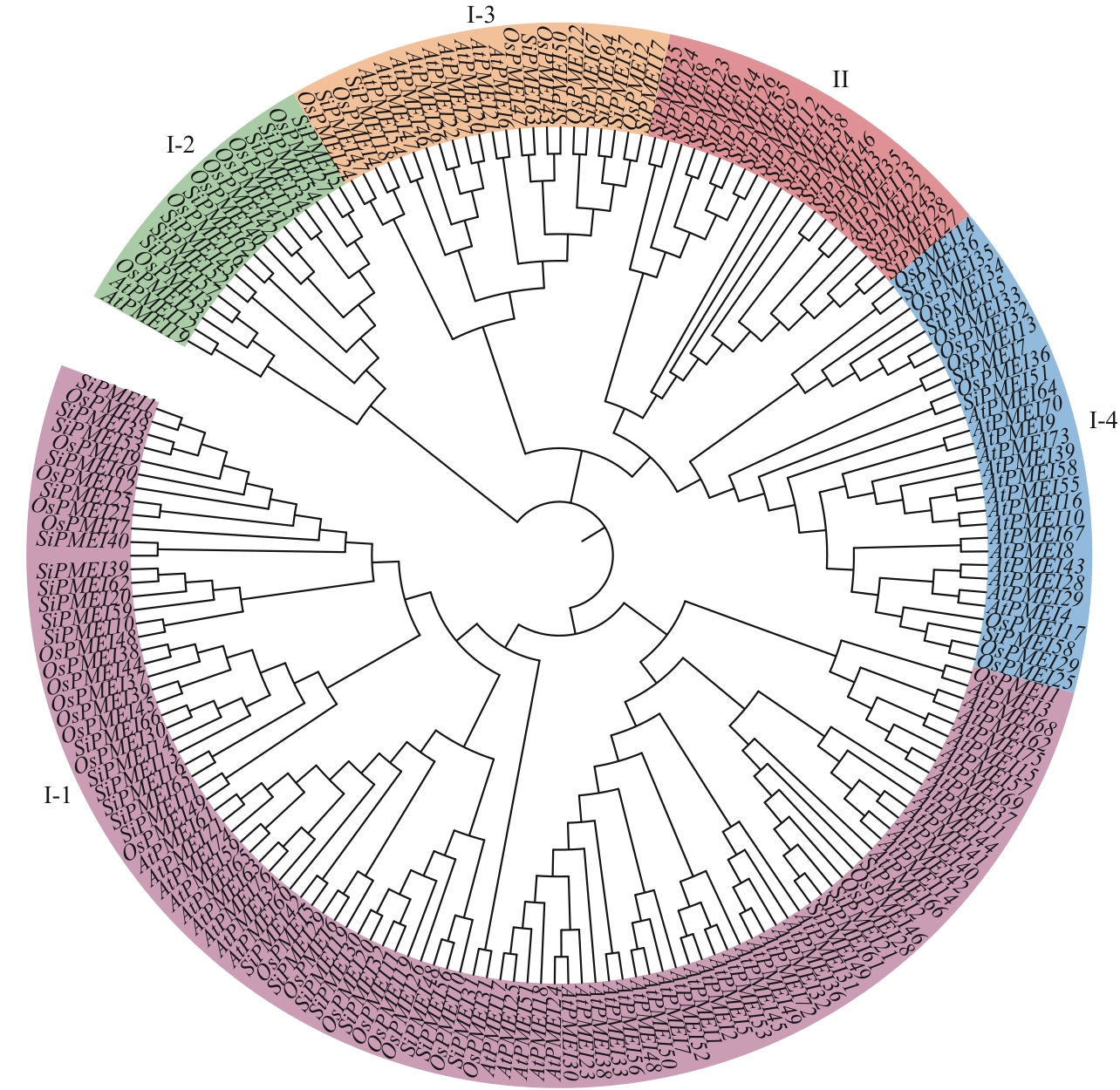
图2 谷子(Si)、水稻(Os)、拟南芥(At)PMEI基因家族系统发育进化树
Fig. 2 Phylogenetic evolutionary tree of the PMEI gene family in Setaria italica (Si), Oryza sativa (Os), and Arabidopsis (At)
重复基因1 Duplicated gene l | 重复基因2 Duplicated gene 2 | 非同义替换率 Ka | 同义替换率 Ks | 比值 Ka/Ks |
|---|---|---|---|---|
| SiPMEI46 | SiPMEI41 | 0.586 044 760 | 0.756 527 598 | 0.774 650 867 |
| SiPMEI29 | SiPMEI19 | 0.310 390 814 | 0.409 962 679 | 0.757 119 686 |
| SiPMEI46 | SiPMEI27 | 0.484 690 326 | 0.680 501 629 | 0.712 254 469 |
| SiPMEI32 | SiPMEI30 | 0.404 813 285 | 0.573 874 330 | 0.705 404 065 |
| SiPMEI26 | SiPMEI27 | 0.591 358 520 | 0.538 368 419 | 1.098 427 208 |
| SiPMEI6 | SiPMEI23 | 0.202 922 616 | 0.397 601 411 | 0.510 366 941 |
| SiPMEI5 | SiPMEI34 | 0.660 998 002 | 0.498 090 476 | 1.327 064 124 |
| SiPMEI63 | SiPMEI50 | 0.617 173 087 | 0.439 225 359 | 1.405 139 924 |
表2 SiPMEI基因进化中复制事件的选择压力参数
Table 2 Selection pressure parameters for replication events in the evolution of SiPMEI genes
重复基因1 Duplicated gene l | 重复基因2 Duplicated gene 2 | 非同义替换率 Ka | 同义替换率 Ks | 比值 Ka/Ks |
|---|---|---|---|---|
| SiPMEI46 | SiPMEI41 | 0.586 044 760 | 0.756 527 598 | 0.774 650 867 |
| SiPMEI29 | SiPMEI19 | 0.310 390 814 | 0.409 962 679 | 0.757 119 686 |
| SiPMEI46 | SiPMEI27 | 0.484 690 326 | 0.680 501 629 | 0.712 254 469 |
| SiPMEI32 | SiPMEI30 | 0.404 813 285 | 0.573 874 330 | 0.705 404 065 |
| SiPMEI26 | SiPMEI27 | 0.591 358 520 | 0.538 368 419 | 1.098 427 208 |
| SiPMEI6 | SiPMEI23 | 0.202 922 616 | 0.397 601 411 | 0.510 366 941 |
| SiPMEI5 | SiPMEI34 | 0.660 998 002 | 0.498 090 476 | 1.327 064 124 |
| SiPMEI63 | SiPMEI50 | 0.617 173 087 | 0.439 225 359 | 1.405 139 924 |

图4 谷子种内PMEI基因家族共线性分析1-9 代表谷子第1-9染色体;染色体内部以基因密度填充,颜色由蓝到红代表基因密度由小到大
Fig. 4 Collinearity analysis of intra-specific PMEI gene families in foxtail millet1-9 indicates chromosome 1-9 in foxtail millet respectively. The interior of the chromosome is filled with gene density, with colors ranging from blue to red indicating a gradual increase in gene density
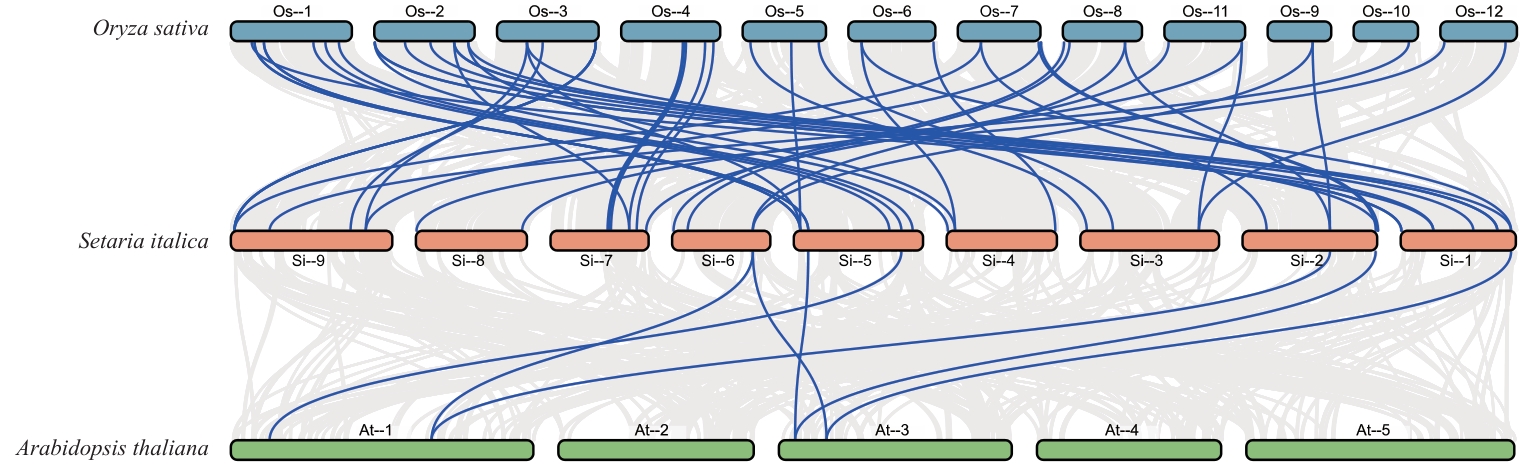
图5 SiPMEI基因家族成员与水稻和拟南芥PMEI基因家族种间共线性分析
Fig. 5 Analysis of inter-specific collinearity between SiPMEI gene family members and PMEI gene families of rice and Arabidopsis
基因名称 Gene name | 低温响应元件 Low-temperature responsive element | 脱落酸响应元件 ABA responsive element | 茉莉酸甲酯响应元件 MeJA responsive element | 干旱响应元件 Drought responsive element | 亚细胞定位 Subcellular location | 结构域 Structural domain |
|---|---|---|---|---|---|---|
| SiPMEI22 | 1 | 8 | 6 | 0 | Chloroplast | Pectinesteras + PMEI |
| SiPMEI30 | 0 | 6 | 10 | 0 | Cell wall | PMEI |
| SiPMEI31 | 0 | 4 | 12 | 0 | Chloroplast | PMEI |
| SiPMEI32 | 2 | 0 | 0 | 4 | Cell wall | PMEI |
| SiPMEI36 | 1 | 4 | 10 | 0 | Cell wall | PMEI |
| SiPMEI38 | 0 | 1 | 4 | 2 | Chloroplast | Pectinesteras + PMEI |
| SiPMEI47 | 1 | 10 | 16 | 1 | Chloroplast | Pectinesteras + PMEI |
| SiPMEI63 | 0 | 7 | 0 | 2 | Cell wall | PMEI |
表3 所选引物及胁迫响应元件的数目
Table 3 Number of selected primers and stress-responsive elements
基因名称 Gene name | 低温响应元件 Low-temperature responsive element | 脱落酸响应元件 ABA responsive element | 茉莉酸甲酯响应元件 MeJA responsive element | 干旱响应元件 Drought responsive element | 亚细胞定位 Subcellular location | 结构域 Structural domain |
|---|---|---|---|---|---|---|
| SiPMEI22 | 1 | 8 | 6 | 0 | Chloroplast | Pectinesteras + PMEI |
| SiPMEI30 | 0 | 6 | 10 | 0 | Cell wall | PMEI |
| SiPMEI31 | 0 | 4 | 12 | 0 | Chloroplast | PMEI |
| SiPMEI32 | 2 | 0 | 0 | 4 | Cell wall | PMEI |
| SiPMEI36 | 1 | 4 | 10 | 0 | Cell wall | PMEI |
| SiPMEI38 | 0 | 1 | 4 | 2 | Chloroplast | Pectinesteras + PMEI |
| SiPMEI47 | 1 | 10 | 16 | 1 | Chloroplast | Pectinesteras + PMEI |
| SiPMEI63 | 0 | 7 | 0 | 2 | Cell wall | PMEI |
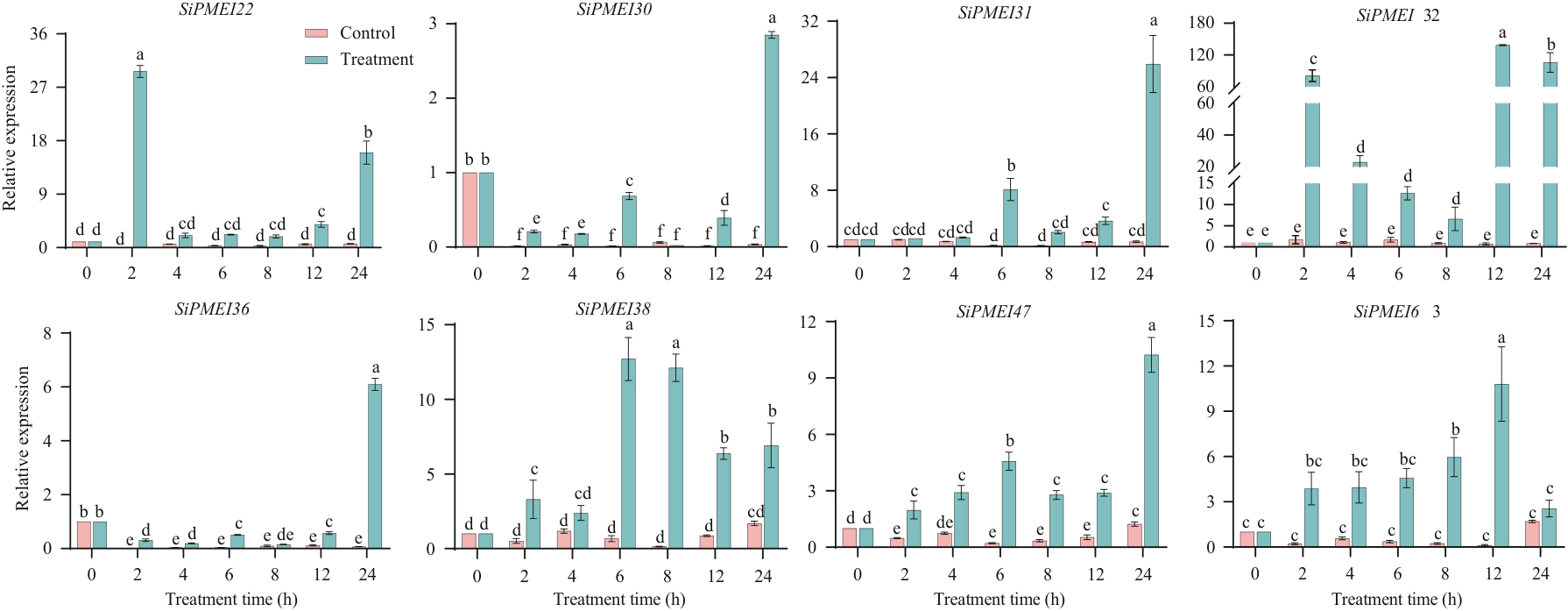
图8 低温胁迫下不同基因的表达水平误差线为标准误,图中不同字母表示差异水平显著(P < 0.05),下同
Fig. 8 Expressions of different genes under low-temperature stressThe error line is the standard error, and the different letters in the figure indicate significant difference levels (P <0.05), the same below
| [1] | 刁现民. 育种创新造就谷子种业新发展 [J]. 中国种业, 2022(4): 4-7. |
| Diao XM. Breeding innovation creates new development of millet seed industry [J]. China Seed Ind, 2022(4): 4-7. | |
| [2] | 周雪, 苏乐平, 李星星, 等. 25个谷子品种(系)主要农艺性状分析及评价 [J]. 中国种业, 2025(2): 95-100, 108. |
| Zhou X, Su LP, Li XX, et al. Analysis and evaluation of main agronomic traits of 25 millet varieties(lines) [J]. China Seed Ind, 2025(2): 95-100, 108. | |
| [3] | 侯保俊, 何太, 杨红霞. 晋北高寒冷凉区谷子地膜覆盖栽培技术 [J]. 农业技术与装备, 2009(5): 34-35. |
| Hou BJ, He T, Yang HX. Cultivation technology of millet in cold area of northern Shanxi [J]. Agric Technol Equip, 2009(5): 34-35. | |
| [4] | 张晓霞. 三种非生物胁迫下谷子qRT-PCR内参基因的筛选与鉴定 [D]. 太谷: 山西农业大学, 2023. |
| Zhang XX. Screening and identification of qRT-PCR reference genes in foxtail millet under threeabiotic stresses [D]. Taigu: Shanxi Agricultural University, 2023. | |
| [5] | 汪明滔, 刘建伟, 赵春钊. 植物调控盐胁迫下细胞壁完整性的分子机制 [J]. 生物技术通报, 2023, 39(11): 18-27. |
| Wang MT, Liu JW, Zhao CZ. Molecular mechanisms of cell wall integrity in plants under salt stress [J]. Biotechnol Bull, 2023, 39(11): 18-27. | |
| [6] | 关月, 费雅婷, 朱祝军, 茹磊. 果胶甲酯酶抑制子在植物生长发育和逆境胁迫中的作用 [J/OL]. 分子植物育种, 2024. . |
| Guan Y, Fei YT, Zhu ZJ, Ru L. The role of pectin methylesterase inhibitors in plant growth, development, and stress response [J/OL]. Mol Plant Breed, 2024. . | |
| [35] | Du XF, Wang ZL, Han KN, et al. Identification and analysis of RNA editing sites of chloroplast genes in foxtail millet [Setaria italica(L.) P.Beauv [J]. Acta Agron Sin, 2022, 48(4): 873-885. |
| [7] | Hothorn M, Wolf S, Aloy P, et al. Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins [J]. Plant Cell, 2004, 16(12): 3437-3447. |
| [8] | 熊媛. 毛果杨PMEI家族分子特征和功能性评估 [D]. 南京: 南京林业大学, 2023. |
| Xiong Y. Molecular characteristics and functional evaluation of PMEI family of Populus tomentosa [D]. Nanjing: Nanjing Forestry University, 2023. | |
| [9] | Balestrieri C, Castaldo D, Giovane A, et al. A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis) [J]. Eur J Biochem, 1990, 193(1): 183-187. |
| [10] | Wang JJ, Ling L, Cai H, et al. Gene-wide identification and expression analysis of the PMEI family genes in soybean (Glycine max) [J]. 3 Biotech, 2020, 10(8): 335. |
| [11] | Liu TT, Yu H, Xiong XP, et al. Genome-wide identification and characterization of pectin methylesterase inhibitor genes in Brassica oleracea [J]. Int J Mol Sci, 2018, 19(11): 3338. |
| [12] | 王文玉, 施旭, 施琪, 等. 水稻PMEI基因家族的全基因组鉴定与生物信息学分析 [J]. 分子植物育种, 2023, 21(23): 7657-7666. |
| Wang WY, Shi X, Shi Q, et al. Genome-wide identification and bioinformatics analysis of PMEI gene family in rice [J]. Mol Plant Breed, 2023, 21(23): 7657-7666. | |
| [13] | Pinzón-Latorre D, Deyholos MK. Characterization and transcript profiling of the pectin methylesterase (PME) and pectin methylesterase inhibitor (PMEI) gene families in flax (Linum usitatissimum) [J]. BMC Genomics, 2013, 14: 742. |
| [14] | Liu NN, Sun Y, Pei YK, et al. A pectin methylesterase inhibitor enhances resistance to Verticillium wilt [J]. Plant Physiol, 2018, 176(3): 2202-2220. |
| [15] | 杨帅, 高尚珠, 卢晗, 等. 植物细胞壁形成及在非生物胁迫中的作用 [J]. 植物生理学报, 2023, 59(7): 1251-1264. |
| Yang S, Gao SZ, Lu H, et al. Plant cell wall development and its function in abiotic stress [J]. Plant Physiol J, 2023, 59(7): 1251-1264. | |
| [16] | 仇婷婷. 木薯果胶甲基酯酶抑制因子MePMEI1功能分析及其互作蛋白的筛选 [D]. 海口: 海南大学, 2020. |
| Qiu TT. Functional analysis of cassava pectin methyl esterase inhibitor MePMEI1 and screening of its interacting proteins [D]. Haikou: Hainan University, 2020. | |
| [17] | 王跃腾. PMEI抑制PME的机理及食品应用型PMEI的改造研究 [D]. 成都: 成都大学, 2023. |
| Wang YT. Mechanism of PMEI inhibiting PME and modification of food application PMEI [D]. Chengdu: Chengdu University, 2023. | |
| [18] | 楚刘阳. 甘蓝型油菜PMEI和XTH基因家族的鉴定与转录谱分析 [D]. 武汉: 华中农业大学, 2021. |
| Chu LY. Identification and transcription analysis of PMEI and XTH gene families in Brassica napus L [D]. Wuhan: Huazhong Agricultural University, 2021. | |
| [19] | Bosch M, Hepler PK. Pectin methylesterases and pectin dynamics in pollen tubes [J]. Plant Cell, 2005, 17(12): 3219-3226. |
| [20] | 盖芸芸, 何婵, 李娜, 等. 超声波预处理对贮藏过程中猕猴桃质构、果胶特性及果胶酶活性的影响 [J]. 食品科技, 2024, 49(5): 26-34. |
| Gai YY, He C, Li N, et al. Effects of ultrasonic pretreatment on texture, pectin properties, and pectinase activity of kiwifruit during storage [J]. Food Sci Technol, 2024, 49(5): 26-34. | |
| [21] | Wachsman G, Zhang JY, Moreno-Risueno MA, et al. Cell wall remodeling and vesicle trafficking mediate the root clock in Arabidopsis [J]. Science, 2020, 370(6518): 819-823. |
| [22] | Zhang GY, Feng J, Wu J, et al. BoPMEI1, a pollen-specific pectin methylesterase inhibitor, has an essential role in pollen tube growth [J]. Planta, 2010, 231(6): 1323-1334. |
| [23] | Coculo D, Lionetti V. The plant invertase/pectin methylesterase inhibitor superfamily [J]. Front Plant Sci, 2022, 13: 863892. |
| [24] | Nguyen HP, Jeong HY, Jeon SH, et al. Rice pectin methylesterase inhibitor28 (OsPMEI28) encodes a functional PMEI and its overexpression results in a dwarf phenotype through increased pectin methylesterification levels [J]. J Plant Physiol, 2017, 208: 17-25. |
| [25] | Müller K, Levesque-Tremblay G, Fernandes A, et al. Overexpression of a pectin methylesterase inhibitor in Arabidopsis thaliana leads to altered growth morphology of the stem and defective organ separation [J]. Plant Signal Behav, 2013, 8(12): e26464. |
| [26] | Peaucelle A, Louvet R, Johansen JN, et al. Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins [J]. Curr Biol, 2008, 18(24): 1943-1948. |
| [27] | An SH, Sohn KH, Choi HW, et al. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance [J]. Planta, 2008, 228(1): 61-78. |
| [28] | Tan C, Liu ZY, Huang SN, et al. Pectin methylesterase inhibitor (PMEI) family can be related to male sterility in Chinese cabbage (Brassica rapa ssp. pekinensis) [J]. Mol Genet Genomics, 2018, 293(2): 343-357. |
| [29] | Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology [J]. Nat Rev Mol Cell Biol, 2009, 10(6): 385-397. |
| [30] | Zhang JZ. Evolution by gene duplication: an update [J]. Trends Ecol Evol, 2003, 18(6): 292-298. |
| [31] | Jeong HY, Nguyen HP, Eom SH, et al. Integrative analysis of pectin methylesterase (PME) and PME inhibitors in tomato (Solanum lycopersicum): Identification, tissue-specific expression, and biochemical characterization [J]. Plant Physiol Biochem, 2018, 132: 557-565. |
| [32] | 廉雪菲. 柑橘果实囊衣发育形成规律研究 [D]. 长沙: 湖南农业大学, 2022. |
| Lian XF. Study on the development and formation law of citrus fruit capsule [D]. Changsha: Hunan Agricultural University, 2022. | |
| [33] | Hong MJ, Kim DY, Lee TG, et al. Functional characterization of pectin methylesterase inhibitor (PMEI) in wheat [J]. Genes Genet Syst, 2010, 85(2): 97-106. |
| [34] | 陈逸飞, 王文博, 王赛赛, 祝建波. 甜瓜PMEI基因家族的鉴定和表达分析 [J/OL]. 分子植物育种, 2023. . |
| Chen YF, Wang WB, Wang SS, Zhu JB. Identification and expression analysis of PMEI gene family in melon [J/OL]. Mol Plant Breed, 2023. . | |
| [35] | 杜晓芬, 王智兰, 韩康妮, 等. 谷子叶绿体基因RNA编辑位点的鉴定与分析 [J]. 作物学报, 2022, 48(4): 873-885. |
| [1] | 林佳怡, 陈强, 张磊, 刘宏鑫, 郑晓明, 逄洪波. 褪黑素在植物低温胁迫中的研究进展[J]. 生物技术通报, 2025, 41(7): 37-48. |
| [2] | 张泽, 杨秀丽, 宁东贤. 花生4CL基因家族鉴定及对干旱与盐胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 117-127. |
| [3] | 程珊, 王会伟, 陈晨, 朱雅婧, 李春鑫, 别海, 王树峰, 陈献功, 张向歌. 油莎豆MYB转录因子基因CeMYB154克隆及耐盐功能分析[J]. 生物技术通报, 2025, 41(6): 218-228. |
| [4] | 王苗苗, 赵相龙, 王召明, 刘志鹏, 闫龙凤. 花苜蓿TCP基因家族的鉴定及其在干旱胁迫下的表达模式分析[J]. 生物技术通报, 2025, 41(6): 179-190. |
| [5] | 许慧珍, SHANTWANA Ghimire, RAJU Kharel, 岳云, 司怀军, 唐勋. 马铃薯SUMO E3连接酶基因家族分析及StSIZ1基因的克隆与表达模式分析[J]. 生物技术通报, 2025, 41(6): 119-129. |
| [6] | 宗建伟, 邓海芳, 蔡沅原, 常雅雯, 朱雅琦, 杨雨华. AM真菌对干旱胁迫下文冠果根系形态和叶片结构耦合的影响[J]. 生物技术通报, 2025, 41(6): 167-178. |
| [7] | 何卫, 李俊怡, 李新妮, 马雪华, 邢媛, 曹晓宁, 乔治军, 刘思辰. 谷子泛素连接酶U-box E3基因家族的鉴定及响应非生物胁迫分析[J]. 生物技术通报, 2025, 41(5): 104-118. |
| [8] | 杨春, 王晓倩, 王红军, 晁跃辉. 蒺藜苜蓿MtZHD4基因克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2025, 41(5): 244-254. |
| [9] | 田琴, 刘奎, 吴翔纬, 纪媛媛, 曹一博, 张凌云. 转录因子VcMYB17调控蓝莓抗旱性的功能研究[J]. 生物技术通报, 2025, 41(4): 198-210. |
| [10] | 班秋艳, 赵鑫月, 迟文静, 黎俊生, 王琼, 夏瑶, 梁丽云, 贺巍, 李叶云, 赵广山. 茶树光敏色素互作因子CsPIF3a的克隆及其与光温逆境的响应[J]. 生物技术通报, 2025, 41(4): 256-265. |
| [11] | 卢勇杰, 夏海乾, 李永铃, 张文建, 余婧, 赵会纳, 王兵, 许本波, 雷波. 烟草AP2/ERF转录因子NtESR2的克隆及功能分析[J]. 生物技术通报, 2025, 41(4): 266-277. |
| [12] | 钱祺, 王增辉, 孙荣华, 罗英智, 苏良辰. 花生蛋白磷酸酶AhPDCP37的抗旱性功能研究[J]. 生物技术通报, 2025, 41(3): 98-103. |
| [13] | 刘洁, 王飞, 陶婷, 张玉静, 陈浩婷, 张瑞星, 石玉, 张毅. 过表达SlWRKY41提高番茄幼苗抗旱性[J]. 生物技术通报, 2025, 41(2): 107-118. |
| [14] | 李禹欣, 李苗, 杜晓芬, 韩康妮, 连世超, 王军. 谷子SiSAP基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(1): 143-156. |
| [15] | 孔青洋, 张晓龙, 李娜, 张晨洁, 张雪云, 于超, 张启翔, 罗乐. 单叶蔷薇GRAS转录因子家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 210-220. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||