








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 256-264.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0122
• 研究报告 • 上一篇
收稿日期:2025-02-06
出版日期:2025-09-26
发布日期:2025-09-24
通讯作者:
陈仲,男,博士,教授,研究方向 :林木功能基因组学;E-mail: zhongchen@bjfu.edu.cn作者简介:张超超,男,硕士研究生,研究方向 :树木分子生物学;E-mail: zhangcc99@bjfu.edu.cn
基金资助:
ZHANG Chao-chao( ), HAN Kai-yuan, WANG Tong, CHEN Zhong(
), HAN Kai-yuan, WANG Tong, CHEN Zhong( )
)
Received:2025-02-06
Published:2025-09-26
Online:2025-09-24
摘要:
目的 探究毛白杨PtoYABBY2和PtoYABBY12在开花和叶片发育过程中的作用,为毛白杨分子设计育种提供有价值的参考基因。 方法 利用同源克隆法克隆毛白杨PtoYABBY2和PtoYABBY12,并对其进行生物信息学、系统发育、表达模式、亚细胞定位分析,通过拟南芥异源转化探究这2个基因的功能。 结果 毛白杨PtoYABBY2和PtoYABBY12具有完整的YABBY结构域和C2C2结构域,二者属于YABBY基因家族YAB2亚类的基因成员,蛋白均定位于细胞核中。PtoYABBY2和PtoYABBY12在毛白杨雌雄花芽发育过程中呈现不同的表达规律;在毛白杨根、茎、幼叶和成熟叶片中均有表达,且在成熟叶片中表达量最高。与野生型拟南芥相比,PtoYABBY2和PtoYABBY12的过表达植株均表现出明显的早花表型,但叶片形态、花序结构和花器官形态未发生显著变化。过表达转基因株系中,促进开花基因AtFT、AtSOC1、AtFUL和AtLFY相对表达量高于野生型拟南芥,抑制开花基因AtFLC相对表达量显著低于野生型拟南芥。 结论 毛白杨PtoYABBY2和PtoYABBY12在开花过程中发挥关键作用,可正向促进植物提前开花。
张超超, 韩开元, 王彤, 陈仲. 毛白杨PtoYABBY2和PtoYABBY12的克隆及功能分析[J]. 生物技术通报, 2025, 41(9): 256-264.
ZHANG Chao-chao, HAN Kai-yuan, WANG Tong, CHEN Zhong. Cloning and Functional Analysis of PtoYABBY2 and PtoYABBY12 in Populus tomentosa[J]. Biotechnology Bulletin, 2025, 41(9): 256-264.
引物名称 Primer name | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) | 用途 Purpose |
|---|---|---|---|
| PtoYAB2 | ATGCATCACATAAACGAAAGGAAA | TCACAGGGAGCCATTGATCTCGTA | 扩增PCR |
| PtoYAB12 | ATGTCTCTAGACATTGCTTCTGAACG | TTATGCATCCATGCCGGTG | |
| PtoYAB2-qPCR | CTTTGGACTGAAGCTGGACA | TCACAGGGAGCCATTGAT | RT-qPCR |
| PtoYAB12-qPCR | TCCCAGTCTTCTTCCTCGGG | GTTCTTGGCTGCATTGCTGA | |
| PtACTIN | TATCTCCTCTGTCTCCGACT | GCATCATCACCTGCAAAC | |
| AtFT (AT1G65480) | CTTGGCAGGCAAACAGTGTATGCAC | GCCACTCTCCCTCTGACAATTGTAGA | |
| AtFUL (AT5G60910) | TTGCAAGATCACAACAATTCGCTTCT | GAGAGTTTGGTTCCGTCAACGACGAT | |
| AtSOC1 (AT2G45660) | AGCTGCAGAAAACGAGAAGCTCTCTG | GGGCTACTCTCTTCATCACCTCTTCC | |
| AtLFY (AT5G61850) | ACGCCGTCATTTGCTACTCT | CTTTCTCCGTCTCTGCTGCT | |
| AtFLC (AT5G10140) | CTAGCCAGATGGAGAATAATCATCATG | TTAAGGTGGCTAATTAAGTAGTGGGAG | |
| AtACTIN2 (AT3G18780) | AGTGGTCGTACAACCGGTATTGT | GATGGCATGGAGGAAGAGAGAAAC |
表1 引物序列
Table 1 Primer sequence
引物名称 Primer name | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) | 用途 Purpose |
|---|---|---|---|
| PtoYAB2 | ATGCATCACATAAACGAAAGGAAA | TCACAGGGAGCCATTGATCTCGTA | 扩增PCR |
| PtoYAB12 | ATGTCTCTAGACATTGCTTCTGAACG | TTATGCATCCATGCCGGTG | |
| PtoYAB2-qPCR | CTTTGGACTGAAGCTGGACA | TCACAGGGAGCCATTGAT | RT-qPCR |
| PtoYAB12-qPCR | TCCCAGTCTTCTTCCTCGGG | GTTCTTGGCTGCATTGCTGA | |
| PtACTIN | TATCTCCTCTGTCTCCGACT | GCATCATCACCTGCAAAC | |
| AtFT (AT1G65480) | CTTGGCAGGCAAACAGTGTATGCAC | GCCACTCTCCCTCTGACAATTGTAGA | |
| AtFUL (AT5G60910) | TTGCAAGATCACAACAATTCGCTTCT | GAGAGTTTGGTTCCGTCAACGACGAT | |
| AtSOC1 (AT2G45660) | AGCTGCAGAAAACGAGAAGCTCTCTG | GGGCTACTCTCTTCATCACCTCTTCC | |
| AtLFY (AT5G61850) | ACGCCGTCATTTGCTACTCT | CTTTCTCCGTCTCTGCTGCT | |
| AtFLC (AT5G10140) | CTAGCCAGATGGAGAATAATCATCATG | TTAAGGTGGCTAATTAAGTAGTGGGAG | |
| AtACTIN2 (AT3G18780) | AGTGGTCGTACAACCGGTATTGT | GATGGCATGGAGGAAGAGAGAAAC |
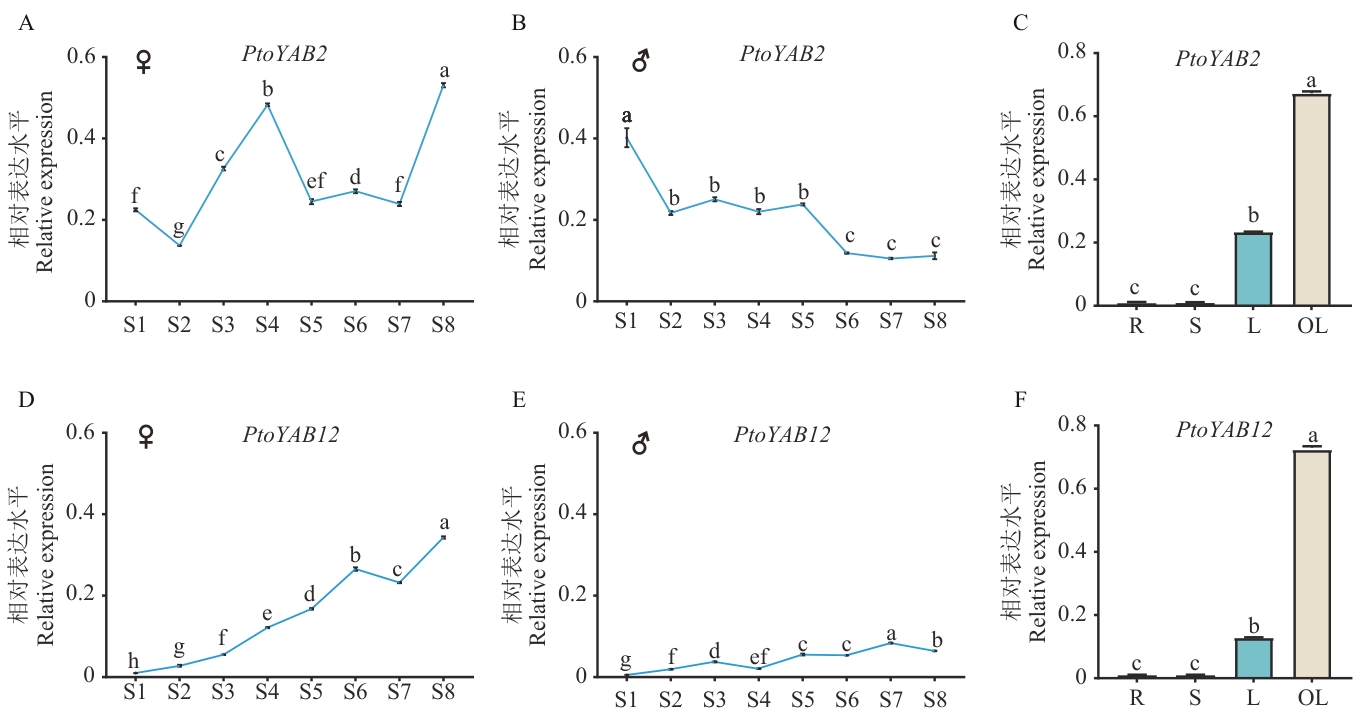
图5 PtoYAB2和PtoYAB12在毛白杨雌雄花芽和不同组织中的表达规律S1‒S8代表8个关键发育时期(S1成花诱导期、S2花原基形成期、S3器官发生期、S4伸长期、S5孢子形成期、S6和S7休眠期、S8孢子发生期)的花芽;R、S、L、OL分别代表根、茎、幼嫩叶片和成熟叶片;♀表示雌花芽,♂表示雄花芽。不同小写字母表示在0.05水平上差异显著,下同
Fig. 5 Expressions of PtoYAB2 and PtoYAB12 in male and female flower buds and different tissues of P. tomentosaS1‒S8 refer to the flower buds at the eight key developmental stages of flower bud development (S1 flower induction stage, S2 flower primordium formation stage, S3 organogenesis stage, S4 elongation stage, S5 archespore formation stage, S6 and S7 dormancy stage, and S8 archespore genesis stage); R, S, L and OL refer to the root, stem, young leaf and mature leaf respectively. ♀ indicates female flower buds and ♂ indicates male flower buds. Different lowercase letters indicate significant differences at the 0.05 level. The same below
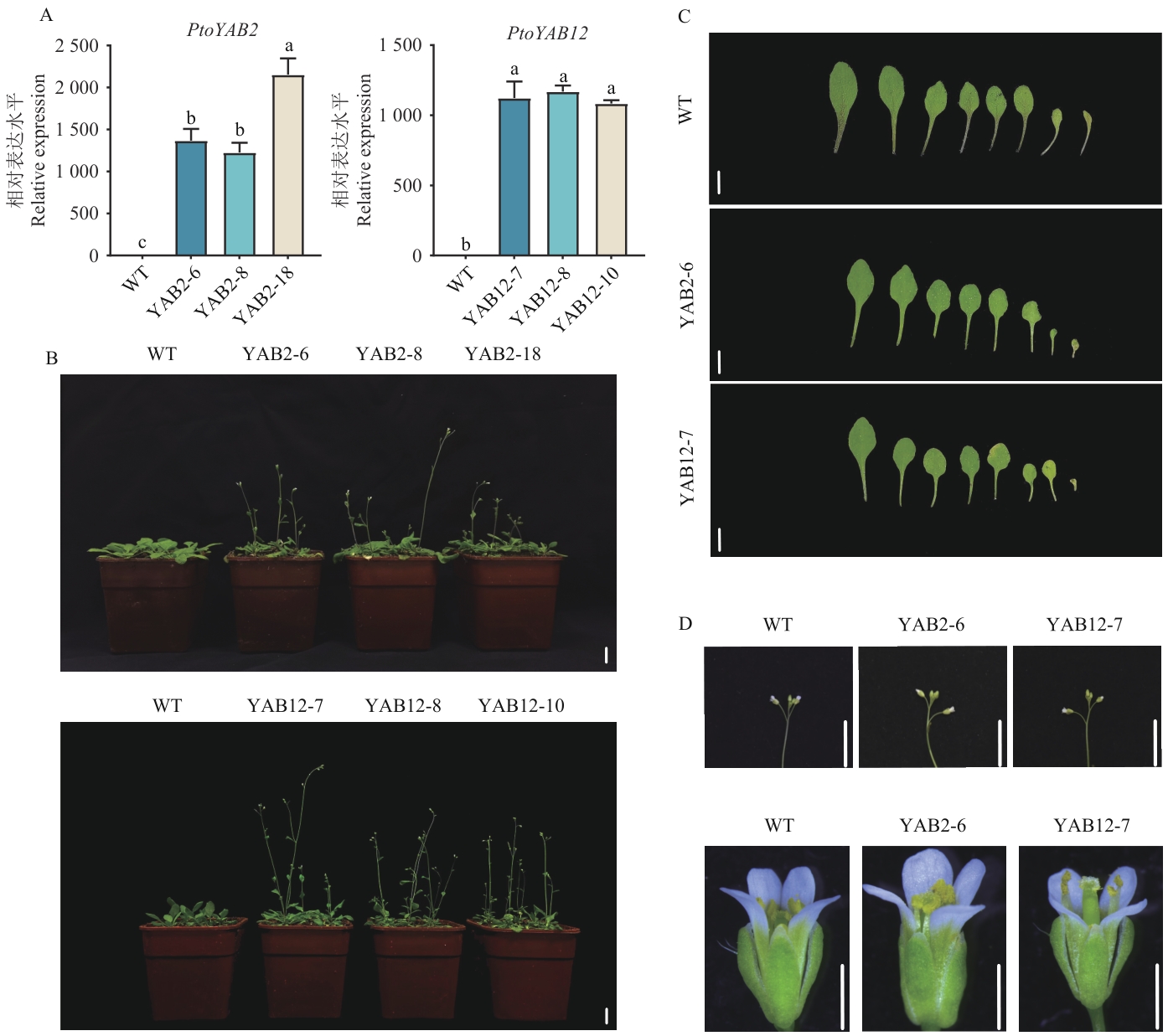
图6 转基因拟南芥鉴定与表型分析A:转基因拟南芥阳性鉴定;B:转基因拟南芥与野生型开花时间差异对比;C:转基因拟南芥与野生型叶片表型对比;D:转基因拟南芥与野生型无限花序和花器官对比。YAB2-6、YAB2-8、YAB2-18为PtoYAB2转基因拟南芥株系,YAB12-7、YAB12-8和YAB12-10为PtoYAB12转基因拟南芥株系,下同。B和C中比例尺均为1 cm,D中比例尺从上至下分别为1 cm和1 mm
Fig. 6 Identification and phenotypic analysis of transgenic A. thalianaA: Positive identification of transgenic Arabidopsis thaliana. B: Comparison of flowering time between transgenic A. thaliana and wild type. C: Comparison of leaf phenotypes between transgenic A. thaliana and wild type. D: Comparison of indeterminate inflorescences and floral organs between transgenic A. thaliana and wild type. YAB2-6, YAB2-8, and YAB2-18 are PtoYAB2 transgenic A. thaliana lines, and YAB12-7, YAB12-8, and YAB12-10 are PtoYAB12 transgenic A. thaliana lines, the same below. The scale of B and C is 1 cm, the scale of D from top to bottom is 1 cm and 1mm
株系 Line | 光照条件 Light condition | 植株数 Number of plants | 开花时间 Time of anthesis (d) |
|---|---|---|---|
| WT (Col) | LD | 10 | 20.33±0.36 |
| YAB2-6 | LD | 11 | 16.00±0.25** |
| YAB2-8 | LD | 12 | 16.50±0.36** |
| YAB2-18 | LD | 10 | 16.00±0.37** |
| YAB12-7 | LD | 12 | 16.16±0.37** |
| YAB12-8 | LD | 12 | 16.00±0.30** |
| YAB12-10 | LD | 10 | 15.75±0.25** |
表2 野生型与转基因拟南芥抽薹开花时间
Table 2 Bolting flowering time of wild-type and transgenic A. thaliana
株系 Line | 光照条件 Light condition | 植株数 Number of plants | 开花时间 Time of anthesis (d) |
|---|---|---|---|
| WT (Col) | LD | 10 | 20.33±0.36 |
| YAB2-6 | LD | 11 | 16.00±0.25** |
| YAB2-8 | LD | 12 | 16.50±0.36** |
| YAB2-18 | LD | 10 | 16.00±0.37** |
| YAB12-7 | LD | 12 | 16.16±0.37** |
| YAB12-8 | LD | 12 | 16.00±0.30** |
| YAB12-10 | LD | 10 | 15.75±0.25** |
| [1] | Finet C, Floyd SK, Conway SJ, et al. Evolution of the YABBY gene family in seed plants [J]. Evol Dev, 2016, 18(2): 116-126. |
| [2] | Han HQ, Liu Y, Jiang MM, et al. Identification and expression analysis of YABBY family genes associated with fruit shape in tomato (Solanum lycopersicum L.) [J]. Genet Mol Res, 2015, 14(2): 7079-7091. |
| [3] | Zhao SP, Lu D, Yu TF, et al. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses [J]. Plant Physiol Biochem, 2017, 119: 132-146. |
| [4] | Bonaccorso O, Lee JE, Puah L, et al. FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor [J]. BMC Plant Biol, 2012, 12: 176. |
| [5] | Bowman JL, Smyth DR. CRABS CLAW a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains [J]. Development, 1999, 126(11): 2387-2396. |
| [6] | Zhang XL, Yang ZP, Zhang J, et al. Ectopic expression of BraYAB1-702, a member of YABBY gene family in Chinese cabbage, causes leaf curling, inhibition of development of shoot apical meristem and flowering stage delaying in Arabidopsis thaliana [J]. Int J Mol Sci, 2013, 14(7): 14872-14891. |
| [7] | Zhang X, Ding L, Song AP, et al. Dwarf and robust plant regulates plant height via modulating gibberellin biosynthesis in Chrysanthemum [J]. Plant Physiol, 2022, 190(4): 2484-2500. |
| [8] | Huang ZJ, Van Houten J, Gonzalez G, et al. Genome-wide identification, phylogeny and expression analysis of SUN OFP and YABBY gene family in tomato [J]. Mol Genet Genomics, 2013, 288(3/4): 111-129. |
| [9] | Yang ZE, Gong Q, Wang LL, et al. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses [J]. Front Genet, 2018, 9: 33. |
| [10] | Han KY, Lai M, Zhao TY, et al. Plant YABBY transcription factors: a review of gene expression, biological functions, and prospects [J]. Crit Rev Biotechnol, 2025, 45(1): 214-235. |
| [11] | Yang H, Shi GX, Li X, et al. Overexpression of a soybean YABBY gene, GmFILa, causes leaf curling in Arabidopsis thaliana [J]. BMC Plant Biol, 2019, 19(1): 234. |
| [12] | Hou HL, Lin Y, Hou XL. Ectopic expression of a pak-choi YABBY gene, BcYAB3, causes leaf curvature and flowering stage delay in Arabidopsis thaliana [J]. Genes, 2020, 11(4): 370. |
| [13] | 郭效诚. 蜡梅YABBY家族CpYABBY5-1与CpCRC功能研究 [D]. 重庆: 西南大学, 2023. |
| Guo XC. Study on CpYABBY5-1 and CpCRC function of Chimonanthus praecox YABBY family [D]. Chongqing: Southwest University, 2023. | |
| [14] | Sun XD, Guan YL, Hu XY. Isolation and characterization of IaYABBY2 gene from Incarvillea arguta [J]. Plant Mol Biol Report, 2014, 32(6): 1219-1227. |
| [15] | Liu SN, Li XF, Yang H, et al. Ectopic expression of BoYAB1, a member of YABBY gene family in Bambusa oldhamii, causes leaf curling and late flowering in Arabidopsis thaliana [J]. J Hortic Sci Biotechnol, 2020, 95(2): 169-174. |
| [16] | Ionescu IA, Møller BL, Sánchez-Pérez R. Chemical control of flowering time [J]. J Exp Bot, 2017, 68(3): 369-382. |
| [17] | Gaudinier A, Blackman BK. Evolutionary processes from the perspective of flowering time diversity [J]. New Phytol, 2020, 225(5): 1883-1898. |
| [18] | Zhang TP, Li CY, Li DX, et al. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants [J]. J Plant Res, 2020, 133(6): 751-763. |
| [19] | Li J, Chen TT, Gao K, et al. Unravelling the novel sex determination genotype with ‘ZY’ and a distinctive 2.15-2.95 Mb inversion among poplar species through haplotype-resolved genome assembly and comparative genomics analysis [J]. Mol Ecol Resour, 2024, 24(7): e14002. |
| [20] | Chen Z, Rao P, Yang XY, et al. A global view of transcriptome dynamics during male floral bud development in Populus tomentosa [J]. Sci Rep, 2018, 8(1): 722. |
| [21] | 郝颖颖, 周明珠, 韩开元, 等. 毛白杨PtYABBY基因家族3个成员的克隆与表达分析 [J]. 北京林业大学学报, 2022, 44(1): 48-57. |
| Hao YY, Zhou MZ, Han KY, et al. Cloning and expression analysis of three members of PtYABBYs in Populus tomentosa [J]. J Beijing For Univ, 2022, 44(1): 48-57. | |
| [22] | Xiang J, Liu RQ, Li TM, et al. Isolation and characterization of two VpYABBY genes from wild Chinese Vitis pseudoreticulata [J]. Protoplasma, 2013, 250(6): 1315-1325. |
| [23] | Jang S, Hur J, Kim SJ, et al. Ectopic expression of OsYAB1 causes extra stamens and carpels in rice [J]. Plant Mol Biol, 2004, 56(1): 133-143. |
| [24] | van der Knaap E, Chakrabarti M, Chu YH, et al. What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape [J]. Front Plant Sci, 2014, 5: 227. |
| [25] | Shi TT, Zhou L, Ye YF, et al. Characterization of YABBY transcription factors in Osmanthus fragrans and functional analysis of OfYABBY12 in floral scent formation and leaf morphology [J]. BMC Plant Biol, 2024, 24(1): 589. |
| [26] | Helliwell CA, Wood CC, Robertson M, et al. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex [J]. Plant J, 2006, 46(2): 183-192. |
| [27] | Shen LS, Zhang Y, Sawettalake N. A Molecular switch for FLOWERING LOCUS C activation determines flowering time in Arabidopsis [J]. Plant Cell, 2022, 34(2): 818-833. |
| [28] | Whittaker C, Dean C. The FLC locus: a platform for discoveries in epigenetics and adaptation [J]. Annu Rev Cell Dev Biol, 2017, 33: 555-575. |
| [29] | Xie YR, Zhou Q, Zhao YP, et al. FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate Arabidopsis flowering [J]. Mol Plant, 2020, 13(3): 483-498. |
| [1] | 史发超, 姜永华, 刘海伦, 文英杰, 严倩. 荔枝LcTFL1基因的克隆与功能分析[J]. 生物技术通报, 2025, 41(9): 159-167. |
| [2] | 刘佳丽, 宋经荣, 赵文宇, 张馨元, 赵子洋, 曹一博, 张凌云. 蓝莓R2R3-MYB基因鉴定及类黄酮调控基因表达分析[J]. 生物技术通报, 2025, 41(9): 124-138. |
| [3] | 张永艳, 郭思健, 李晶, 郝思怡, 李瑞得, 刘嘉鹏, 程春振. 蓝莓花青素相关VcGSTF19基因的克隆及功能研究[J]. 生物技术通报, 2025, 41(9): 139-146. |
| [4] | 李玉珍, 李梦丹, 张蔚, 彭婷. 基于月季扩展蛋白基因家族鉴定的野蔷薇RmEXPB2基因功能研究[J]. 生物技术通报, 2025, 41(9): 182-194. |
| [5] | 黄诗宇, 田姗姗, 杨天为, 高曼熔, 张尚文. 赤苍藤WRI1基因家族的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(8): 242-254. |
| [6] | 康琴, 汪霞, 谌明洋, 徐静天, 陈诗兰, 廖平杨, 许文志, 吴卫, 徐东北. 薄荷UV-B受体基因McUVR8的克隆与表达分析[J]. 生物技术通报, 2025, 41(8): 255-266. |
| [7] | 王芳, 乔帅, 宋伟, 崔鹏娟, 廖安忠, 谭文芳, 杨松涛. 甘薯IbNRT2基因家族全基因组鉴定和表达分析[J]. 生物技术通报, 2025, 41(7): 193-204. |
| [8] | 许慧珍, SHANTWANA Ghimire, RAJU Kharel, 岳云, 司怀军, 唐勋. 马铃薯SUMO E3连接酶基因家族分析及StSIZ1基因的克隆与表达模式分析[J]. 生物技术通报, 2025, 41(6): 119-129. |
| [9] | 彭绍智, 王登科, 张祥, 戴雄泽, 徐昊, 邹学校. 辣椒CaFD1基因克隆、表达特征及功能验证[J]. 生物技术通报, 2025, 41(5): 153-164. |
| [10] | 王天禧, 杨炳松, 潘荣君, 盖文贤, 梁美霞. 苹果PLATZ基因家族鉴定及MdPLATZ9基因功能研究[J]. 生物技术通报, 2025, 41(4): 176-187. |
| [11] | 宋佳怡, 苏芸丽, 郑兴艳, 夏文念, 杨冬梅, 胡慧贞. 金鱼草Expansin基因家族鉴定及其抗核盘菌相关基因筛选[J]. 生物技术通报, 2025, 41(4): 227-242. |
| [12] | 张益瑄, 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰. 马铃薯DIR家族全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(3): 123-136. |
| [13] | 宋姝熠, 蒋开秀, 刘欢艳, 黄亚成, 刘林娅. ‘红阳’猕猴桃TCP基因家族鉴定及其在果实中的表达分析[J]. 生物技术通报, 2025, 41(3): 190-201. |
| [14] | 匡健华, 程志鹏, 赵永晶, 杨洁, 陈润乔, 陈龙清, 胡慧贞. 激素和非生物胁迫下荷花GH3基因家族的表达分析[J]. 生物技术通报, 2025, 41(2): 221-233. |
| [15] | 黄颖, 遇文婧, 刘雪峰, 刁桂萍. 山新杨谷胱甘肽转移酶基因的生物信息学与表达模式分析[J]. 生物技术通报, 2025, 41(2): 248-256. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||