








生物技术通报 ›› 2022, Vol. 38 ›› Issue (11): 185-193.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0237
收稿日期:2022-02-26
出版日期:2022-11-26
发布日期:2022-12-01
作者简介:王博雅,女,博士,讲师,研究方向:植物分子遗传;E-mail:基金资助:
WANG Bo-ya( ), JIANG Yong, HUANG Yan, CAO Ying, HU Shang-lian(
), JIANG Yong, HUANG Yan, CAO Ying, HU Shang-lian( )
)
Received:2022-02-26
Published:2022-11-26
Online:2022-12-01
摘要:
非木造浆是解决国内纸浆短缺的重要手段。慈竹(Bambusa emeiensis)是我国非木造浆的主要原材料之一,提高慈竹中纤维素的含量能够有效提高竹类造浆的效率。通过前期对慈竹进行的转录组测序分析,挖掘出慈竹中一个与植物中纤维素合酶亚基A(cellulose synthase A,CesA)同源的基因,命名为BeCesA4。结果显示,克隆出的BeCesA4基因编码一个含有982个氨基酸的蛋白质,具备CesA家族的保守结构域;BeCesA4在慈竹快速生长的笋与茎中显著表达;过量表达该基因会使转基因植物出现生物量提升、纤维素含量升高和次生细胞壁加厚等现象。结果表明,BeCesA4的表达量与慈竹茎内纤维素的积累呈正相关。本研究结果为进一步揭示慈竹纤维素合成机制奠定了基础。
王博雅, 姜勇, 黄艳, 曹颖, 胡尚连. 慈竹纤维素合酶BeCesA4的克隆及功能分析[J]. 生物技术通报, 2022, 38(11): 185-193.
WANG Bo-ya, JIANG Yong, HUANG Yan, CAO Ying, HU Shang-lian. Cloning and Functional Analysis of BeCesA4 in Bambusa emeiensis[J]. Biotechnology Bulletin, 2022, 38(11): 185-193.
| 引物名称Primer name | 序列Sequence(5'-3') | 用途Usage |
|---|---|---|
| BeCesA4F | TACTACTCGCGATACCCCATAG | 获得BeCesA4基因序列 Obtaining sequence of gene BeCesA4 |
| BeCesA4R | ATACTAACCTACGCCACCTCTC | |
| BeCesA4F(Kpn I) | GGGGTACCCCTACTACTCGCGATACCCCATAG | 构建植物表达载体 Construction of plant expression-vector |
| BeCesA4R(Xba I) | GCTCTAGAGCATACTAACCTACGCCACCTCTC | |
| BeCesA4-RTF | TCACCATCGGCAGCCACCT | 基因表达分析 Analysis of gene expression |
| BeCesA4-RTR | GCTTTTGCAGCAGCCTTTT | |
| Actin-RTF | AAACTGTAATGGTCCTCCCTCCG | 内参基因扩增 Amplification of internal reference |
| Actin-RTR | GCATCATCACAATCACTCTCCGA |
表1 所用引物序列
Table 1 Primer sequence
| 引物名称Primer name | 序列Sequence(5'-3') | 用途Usage |
|---|---|---|
| BeCesA4F | TACTACTCGCGATACCCCATAG | 获得BeCesA4基因序列 Obtaining sequence of gene BeCesA4 |
| BeCesA4R | ATACTAACCTACGCCACCTCTC | |
| BeCesA4F(Kpn I) | GGGGTACCCCTACTACTCGCGATACCCCATAG | 构建植物表达载体 Construction of plant expression-vector |
| BeCesA4R(Xba I) | GCTCTAGAGCATACTAACCTACGCCACCTCTC | |
| BeCesA4-RTF | TCACCATCGGCAGCCACCT | 基因表达分析 Analysis of gene expression |
| BeCesA4-RTR | GCTTTTGCAGCAGCCTTTT | |
| Actin-RTF | AAACTGTAATGGTCCTCCCTCCG | 内参基因扩增 Amplification of internal reference |
| Actin-RTR | GCATCATCACAATCACTCTCCGA |

图2 BeCesA4在进化上与AtCesA8、OsCesA4和BdCe-sA8同源 A:BeCesA4编码序列与其他物种中CesA基因的进化树图。进化树中出现的有拟南芥(Arabidopsis)、水稻(Oryza sativa)、二穗短柄草(Brachy podium distachyon)、杨树(Populus tomentosa)、绿竹(Bambusa oldhamii)以及毛竹(Phyllostachys heterocycla)中鉴定出的CesA家族成员;B:BeCesA4与其他物种CesA蛋白质保守结构域的同源序列比对
Fig.2 BeCesA4 is homologous to AtCesA8,OsCesA4 and BdCesA8 in evolution A:Coding sequence of BeCesA4 and phylogenetic tree with CesA gene in species. The phylogenetic tree there are CesA family members from the species of Arabidopsis,Oryza sativa,Brachy podium distachyon,Populus tomentosa,Bambusa oldhamii and Phyllostachys heterocycla. B:Homologous sequences alignment of BeCesA4 with conserved domain in other species
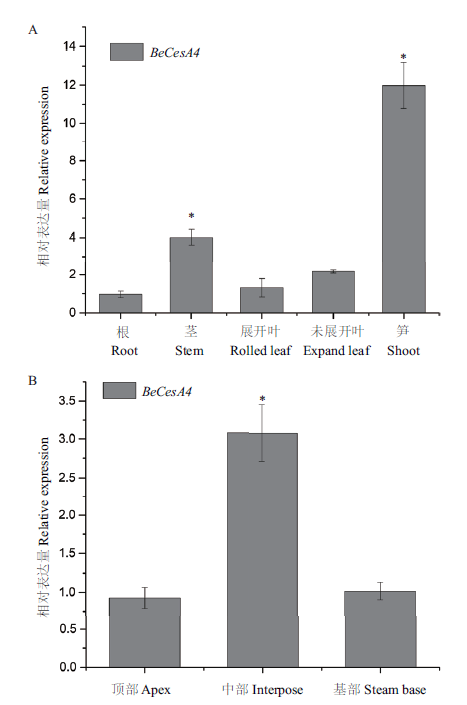
图3 BeCesA4在快速生长部位大量表达 A:慈竹不同部位中BeCesA4的表达量;B:不同高度的笋中BeCesA4的表达量。数据来源于3次独立的生物学重复。“*”号表示根据t检验与其他样本相比有显著性差异(P<0.05),下同
Fig.3 BeCesA4 notably expressed in fast-growing organs A:Expression of BeCesA4 in Bambusa emeiensis organs. B:BeCesA4 expressions differ in shoots at varied heights. Data represent means ± SD from 3 independent replicates. Asterisks indicate significant differences(*P<0.05)according to t test compared with other samples. The same below

图4 BeCesA4转基因毛白杨生物量增加 A:转基因毛白杨中BeCesA4转录水平鉴定;B:BeCesA4转基因毛白杨株高;C:BeCesA4转基因毛白杨生长情况,标尺长度为20 cm
Fig. 4 Biomass increment in BeCesA4 transgenic Populus tomentosa A:Expression analysis of BeCesA4 in transgenic populus. B:Height of transgenic populus and control. C:Growth of transgenic populus,bar length is 20 cm
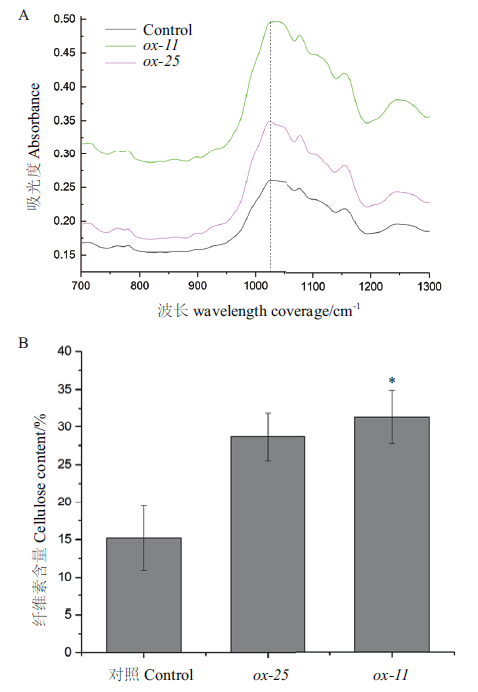
图5 BeCesA4转基因毛白杨纤维素含量增加 A:FITR测定对照与BeCesA4转基因植物中的吸光度;B:对照与BeCesA4转基因植物中纤维素含量的测定
Fig.5 Increase of cellulose content in BeCesA4 transgenic Populus tomentosa A:Absorbance of BeCesA4 transgenic Populus plants and control. B:Cellulose content of BeCesA4 transgenic Populus plants and control
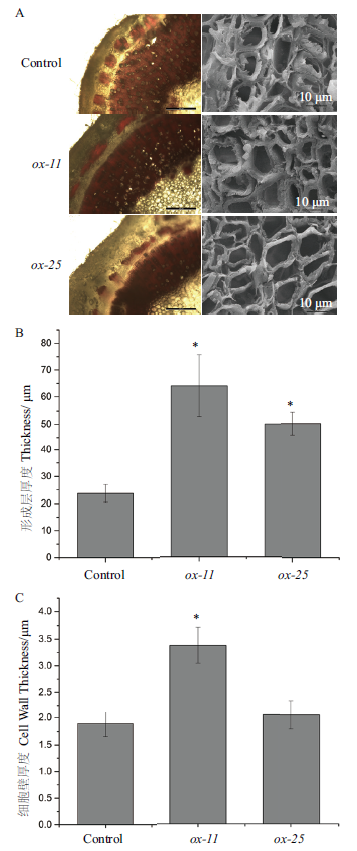
图6 BeCesA4参与细胞壁形成 A-左列:转基因毛白杨茎干切片间苯三酚-盐酸染色染色,A-右列:转基因毛白杨茎干切片扫描电镜照片;B:转基因毛白杨形成层厚度;C:转基因毛白杨细胞壁厚度
Fig.6 BeCesA4 participating in cell wall formation A-left:Phloroglucinol-hydrochloric staining of transgenic Populus tomentosa stem paraffin section. A-right:Scanning electron microscope image of transgenic Populus tomentosa stem paraffin sections. B:Cambium thickness of transgenic Populus tomentosa. C:Cell wall thickness of both transgenic Populus tomentosa and control
| [1] | China Paper Association. 中国造纸协会关于中国造纸工业2002年度报告[J]. 中华纸业, 2003, 24(5):6-9. |
| Association CP. A review on the development of China's paper industry in 2002[J]. China Pulp Pap Ind, 2003, 24(5):6-9. | |
| [2] |
Polko JK, Kieber JJ. The regulation of cellulose biosynthesis in plants[J]. Plant Cell, 2019, 31(2):282-296.
doi: 10.1105/tpc.18.00760 |
| [3] |
McFarlane HE, Döring A, Persson S. The cell biology of cellulose synthesis[J]. Annu Rev Plant Biol, 2014, 65:69-94.
doi: 10.1146/annurev-arplant-050213-040240 pmid: 24579997 |
| [4] | Pérez S, Samain D. Structure and engineering of celluloses[J]. Adv Carbohydr Chem Biochem, 2010, 64:25-116. |
| [5] | 高艳, 陈光辉, 陈秀娟, 等. 植物细胞壁纤维素生物合成的调控[J]. 生物技术通报, 2014(1):1-7. |
| Gao Y, Chen GH, Chen XJ, et al. Regulation of cellulose biosynthesis in plant cell wall[J]. Biotechnol Bull, 2014(1):1-7. | |
| [6] |
Wang LQ, Guo K, Li Y, et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice[J]. BMC Plant Biol, 2010, 10:282.
doi: 10.1186/1471-2229-10-282 URL |
| [7] |
Holland N, Holland D, Helentjaris T, et al. A comparative analysis of the plant cellulose synthase(CesA)gene family[J]. Plant Physiol, 2000, 123(4):1313-1324.
pmid: 10938350 |
| [8] |
Chen ZZ, Hong XH, Zhang HR, et al. Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis[J]. Plant J, 2005, 43(2):273-283.
doi: 10.1111/j.1365-313X.2005.02452.x URL |
| [9] |
Suzuki S, Li LG, Sun YH, et al. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa[J]. Plant Physiol, 2006, 142(3):1233-1245.
pmid: 16950861 |
| [10] |
Zhang SJ, Jiang ZX, Chen J, et al. The cellulose synthase(CesA)gene family in four Gossypium species:phylogenetics, sequence variation and gene expression in relation to fiber quality in Upland cotton[J]. Mol Genet Genomics, 2021, 296(2):355-368.
doi: 10.1007/s00438-020-01758-7 URL |
| [11] |
Xu WJ, Cheng H, Zhu SR, et al. Functional understanding of secondary cell wall cellulose synthases in Populus trichocarpa via the Cas9/gRNA-induced gene knockouts[J]. New Phytol, 2021, 231(4):1478-1495.
doi: 10.1111/nph.17338 URL |
| [12] |
Kumar M, Atanassov I, Turner S. Functional analysis of cellulose synthase(CESA)protein class specificity[J]. Plant Physiol, 2016, 173(2):970-983.
doi: 10.1104/pp.16.01642 URL |
| [13] |
Zhong RQ, Morrison WH 3rd, Freshour GD, et al. Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis[J]. Plant Physiol, 2003, 132(2):786-795.
pmid: 12805608 |
| [14] |
Park S, Ding SY. The N-terminal zinc finger of CELLULOSE SYNTHASE6 is critical in defining its functional properties by determining the level of homodimerization in Arabidopsis[J]. Plant J, 2020, 103(5):1826-1838.
doi: 10.1111/tpj.14870 URL |
| [15] |
Daras G, Templalexis D, Avgeri F, et al. Updating insights into the catalytic domain properties of plant Cellulose synthase(CesA)and Cellulose synthase-like(Csl)proteins[J]. Molecules, 2021, 26(14):4335.
doi: 10.3390/molecules26144335 URL |
| [16] |
Tanaka K, Murata K, Yamazaki M, et al. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall[J]. Plant Physiol, 2003, 133(1):73-83.
pmid: 12970476 |
| [17] |
Li FC, Liu ST, Xu H, et al. A novel FC17/CESA4 mutation causes increased biomass saccharification and lodging resistance by remodeling cell wall in rice[J]. Biotechnol Biofuels, 2018, 11:298.
doi: 10.1186/s13068-018-1298-2 pmid: 30410573 |
| [18] |
Peng ZH, Lu Y, Li LB, et al. The draft genome of the fast-growing non-timber forest species moso bamboo(Phyllostachys heterocycla)[J]. Nat Genet, 2013, 45(4):456-461, 461e1-2.
doi: 10.1038/ng.2569 URL |
| [19] | Zhao HS, Gao ZM, Wang L, et al. Chromosome-level reference genome and alternative splicing atlas of moso bamboo(Phyllostachys edulis)[J]. GigaScience, 2018, 7(10):giy115. |
| [20] |
Guo ZH, Ma PF, Yang GQ, et al. Genome sequences provide insights into the reticulate origin and unique traits of woody bamboos[J]. Mol Plant, 2019, 12(10):1353-1365.
doi: 10.1016/j.molp.2019.05.009 URL |
| [21] |
Chen CY, Hsieh MH, Yang CC, et al. Analysis of the cellulose synthase genes associated with primary cell wall synthesis in Bambusa oldhamii[J]. Phytochemistry, 2010, 71(11/12):1270-1279.
doi: 10.1016/j.phytochem.2010.05.011 URL |
| [22] |
Huang HY, Cheng YS. Heterologous overexpression, purification and functional analysis of plant cellulose synthase from green bamboo[J]. Plant Methods, 2019, 15:80.
doi: 10.1186/s13007-019-0466-0 URL |
| [23] | 陈宇鹏, 曹颖, 胡尚连, 等. 基于高通量测序的慈竹笋转录组分析与基因功能注释[J]. 生物工程学报, 2016, 32(11):1610-1623. |
| Chen YP, Cao Y, Hu SL, et al. Transcriptome analysis and gene function annotation of Bambusa emeiensis shoots based on high-throughput sequencing technology[J]. Chin J Biotechnol, 2016, 32(11):1610-1623. | |
| [24] |
Mazarei M, Baxter HL, Li M, et al. Functional analysis of cellulose synthase CesA4 and CesA6 genes in switchgrass(Panicum virgatum)by overexpression and RNAi-mediated gene silencing[J]. Front Plant Sci, 2018, 9:1114.
doi: 10.3389/fpls.2018.01114 pmid: 30127793 |
| [25] |
Zhang BC, Deng LW, Qian Q, et al. A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice[J]. Plant Mol Biol, 2009, 71(4/5):509-524.
doi: 10.1007/s11103-009-9536-4 URL |
| [26] |
Fan CF, Feng SQ, Huang JF, et al. AtCesA8-driven OsSUS3 expression leads to largely enhanced biomass saccharification and lodging resistance by distinctively altering lignocellulose features in rice[J]. Biotechnol Biofuels, 2017, 10:221.
doi: 10.1186/s13068-017-0911-0 URL |
| [27] |
Li A, Xia T, Xu W, et al. An integrative analysis of four CESA isoforms specific for fiber cellulose production between Gossypium hirsutum and Gossypium barbadense[J]. Planta, 2013, 237(6):1585-1597.
doi: 10.1007/s00425-013-1868-2 URL |
| [28] |
Ye SW, Cai CY, Ren HB, et al. An efficient plant regeneration and transformation system of ma bamboo(Dendrocalamus latiflorus Munro)started from young shoot as explant[J]. Front Plant Sci, 2017, 8:1298.
doi: 10.3389/fpls.2017.01298 URL |
| [29] |
Ye SW, Chen G, Kohnen MV, et al. Robust CRISPR/Cas9 mediated genome editing and its application in manipulating plant height in the first generation of hexaploid Ma bamboo(Dendrocalamus latiflorus Munro)[J]. Plant Biotechnol J, 2020, 18(7):1501-1503.
doi: 10.1111/pbi.13320 URL |
| [30] | Huang BY, Zhuo RY, Fan HJ, et al. An efficient genetic transformation and CRISPR/Cas9-based genome editing system for moso bamboo(Phyllostachys edulis)[J]. Front Plant Sci, 2022, 13:822022. |
| [31] |
Hernández-Blanco C, Feng DX, Hu J, et al. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance[J]. Plant Cell, 2007, 19(3):890-903.
pmid: 17351116 |
| [32] |
Ramírez V, García-Andrade J, Vera P. Enhanced disease resistance to Botrytis cinerea in myb46 Arabidopsis plants is associated to an early down-regulation of CesA genes[J]. Plant Signal Behav, 2011, 6(6):911-913.
doi: 10.4161/psb.6.6.15354 URL |
| [33] |
Coolen S, van Pelt JA, van Wees SCM, et al. Mining the natural genetic variation in Arabidopsis thaliana for adaptation to sequential abiotic and biotic stresses[J]. Planta, 2019, 249(4):1087-1105.
doi: 10.1007/s00425-018-3065-9 URL |
| [34] | 徐梅卿, 戴玉成, 范少辉, 等. 中国竹类病害记述及其病原物分类地位(上)[J]. 林业科学研究, 2006, 19(6):692-699. |
| Xu MQ, Dai YC, Fan SH, et al. Records of bamboo diseases and the taxonomy of their pathogens in China(I)[J]. For Res, 2006, 19(6):692-699. |
| [1] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [2] | 马玉倩, 孙东辉, 岳浩峰, 辛佳瑜, 刘宁, 曹志艳. 具有辅助降解纤维素功能的大斑刚毛座腔菌糖苷水解酶GH61的鉴定、异源表达及功能分析[J]. 生物技术通报, 2023, 39(4): 124-135. |
| [3] | 陈楠楠, 王春来, 蒋振忠, 焦鹏, 关淑艳, 马义勇. 玉米ZmDHN15基因在烟草中的遗传转化及抗冷性分析[J]. 生物技术通报, 2023, 39(4): 259-267. |
| [4] | 牛馨, 张莹, 王茂军, 刘文龙, 路福平, 李玉. 解淀粉芽胞杆菌不同整合位点对外源碱性蛋白酶表达的影响[J]. 生物技术通报, 2022, 38(4): 253-260. |
| [5] | 王玥, 高庆华, 董聪, 罗同阳, 王庆庆. 密码子优化的吡喃糖氧化酶基因在毕赤酵母中的表达[J]. 生物技术通报, 2022, 38(4): 269-277. |
| [6] | 王小桃, 邹杭, 吴怡, 向省维, 吕华, 刘超兰, 林家富, 王欣荣, 褚以文, 宋涛. Paraglaciecola hydrolytica中新型β-琼胶酶Aga2的异源表达及酶学性质分析[J]. 生物技术通报, 2022, 38(11): 258-268. |
| [7] | 张彤彤, 郑登俞, 吴忠义, 张中保, 于荣. 玉米NF-Y转录因子基因ZmNF-YB13响应干旱和盐胁迫的功能分析[J]. 生物技术通报, 2022, 38(10): 115-123. |
| [8] | 廖兆民, 蔡俊, 林建国, 杜馨, 王常高. 黑曲霉葡萄糖氧化酶基因在毕赤酵母中的表达及产酶条件的优化[J]. 生物技术通报, 2021, 37(6): 97-107. |
| [9] | 刘珊, 叶伟, 朱牧孜, 李赛妮, 邓张双, 章卫民. 一种新型酰基转移酶GPAT的克隆、表达与酶学性质研究[J]. 生物技术通报, 2021, 37(11): 257-266. |
| [10] | 赵海燕, 宋晨斌, 刘正亚, 马兴荣, 尚会会, 李安华, 关现军, 王建设. 来源于Laceyella sp.的α-淀粉酶基因克隆、重组表达及酶学性质研究[J]. 生物技术通报, 2020, 36(8): 23-33. |
| [11] | 李卫娜, 申冬玲, 张煜星, 刘学通, 伊日布斯. Mangrovibacterium sp. SH-52耐热内切型海藻酸裂解酶基因的克隆及酶学鉴定[J]. 生物技术通报, 2020, 36(12): 82-90. |
| [12] | 赵江华, 房欢, 张大伟. 微生物次级代谢产物生物合成的研究进展[J]. 生物技术通报, 2020, 36(11): 141-147. |
| [13] | 张钰文, 袁航, 于江悦, 马晓晓, 史超硕, 李玉. 一株高效降解羽毛废弃物菌株的筛选及表达条件优化[J]. 生物技术通报, 2019, 35(9): 93-98. |
| [14] | 陈建军, 刘梁涛, 曹香林. 黄孢原毛平革菌漆酶基因lac1680的克隆、表达及产酶研究[J]. 生物技术通报, 2018, 34(4): 214-220. |
| [15] | 顾源, 寻子琦, 郑菲, 涂涛, 姚斌, 罗会颖. 来源于嗜热真菌Talaromyces leycettanus JCM12802 的类膨胀素基因鉴定及功能探究[J]. 生物技术通报, 2018, 34(10): 172-181. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||