








生物技术通报 ›› 2025, Vol. 41 ›› Issue (5): 82-89.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1246
收稿日期:2024-12-24
出版日期:2025-05-26
发布日期:2025-06-05
通讯作者:
张治国,男,博士,研究员,博士生导师,研究方向 :作物高光效;E-mail: zhangzhiguo@caas.cn作者简介:杜量衡,男,硕士研究生,研究方向 :作物高光效;E-mail: du1150267215@163.com
基金资助:
DU Liang-heng1,2( ), TANG Huang-lei2, ZHANG Zhi-guo2(
), TANG Huang-lei2, ZHANG Zhi-guo2( )
)
Received:2024-12-24
Published:2025-05-26
Online:2025-06-05
摘要:
目的 探究水稻开花期的调控基因与图位克隆,阐明其开花遗传与分子机制,完善水稻抽穗期基因的调控网络,为水稻生产育种提供实践意义。 方法 以水稻长日照条件下延迟开花突变体elm1为材料,统计开花期等农艺性状,通过正反交实验构建群体,统计F2群体表型进行遗传规律分析,与籼稻Dular构建图位克隆群体并进行基因定位,并对精细定位区间内候选基因进行测序,结合生物信息学等手段对候选基因进行预测与分析,使用AlphaFold2软件预测蛋白结构的变化。 结果 elm1突变体在长日照条件下开花时间显著增加,其受一对单隐性核基因控制,图位克隆到ELM1基因,ELM1基因编码SET结构域组蛋白甲基转移酶,等位于已报道的lvp1突变体,测序结果显示在elm1突变体中LOC_Os09g13740基因的第5外显子(ATG下游3 293 bp处)发生点突变(G变为T),导致该突变位点由甘氨酸(亲水)突变为缬氨酸(疏水),AlphaFold2蛋白结构预测表明该突变导致蛋白构象发生改变,对蛋白功能可能有一定的影响。田间试验表明,在合适纬度的区域种植elm1突变体,突变体表现穗粒数明显增多且增产的表型。 结论 elm1突变体的突变位点为一弱等位突变,该等位突变体在适宜的纬度区域下育种可提升水稻产量,研究证明突变体elm1是一份优异的等位变异材料。
杜量衡, 唐黄磊, 张治国. 控制水稻光响应基因ELM1的图位克隆[J]. 生物技术通报, 2025, 41(5): 82-89.
DU Liang-heng, TANG Huang-lei, ZHANG Zhi-guo. Map-based Cloning of Light-responsive Gene ELM1 in Rice[J]. Biotechnology Bulletin, 2025, 41(5): 82-89.
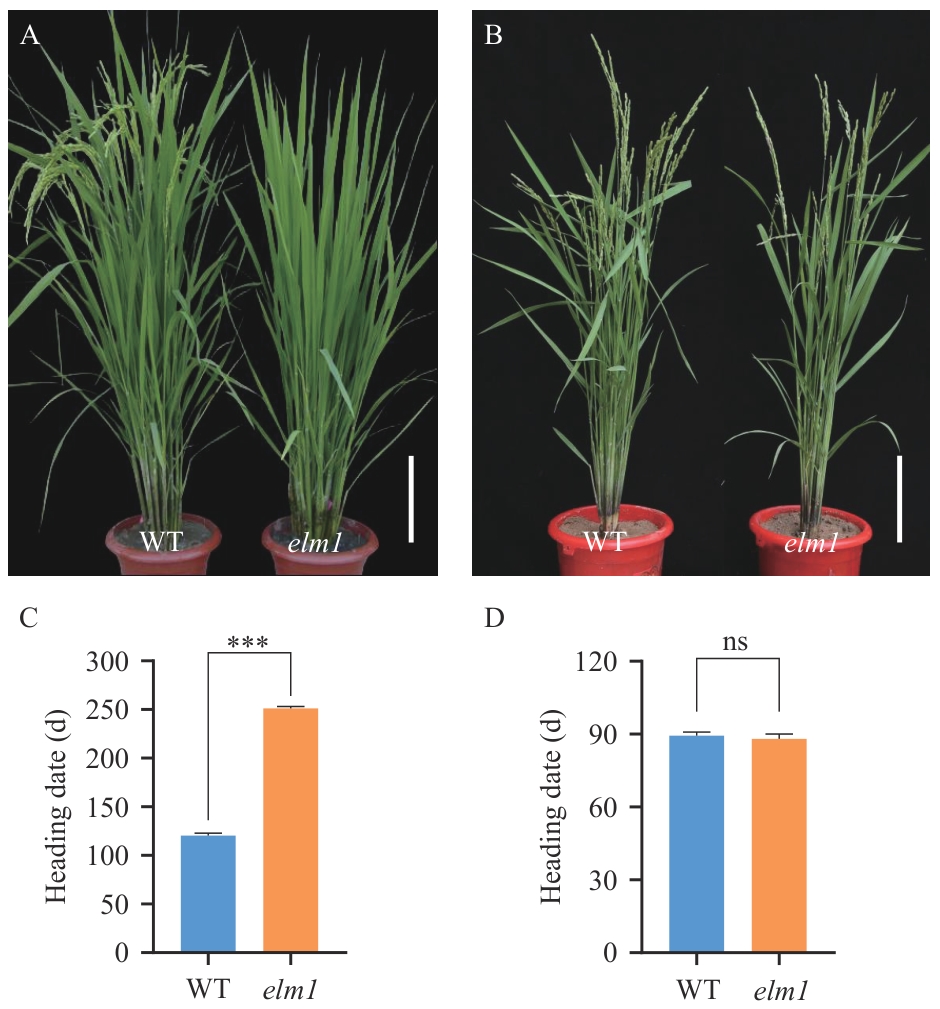
图1 野生型与突变体elm1的开花期表型分析A:野生型(WT)与突变体elm1在北京试验田的开花期植株比较(Bar=20 cm);B:野生型(WT)与突变体elm1的在海南三亚试验田开花期植株比较(Bar=20 cm);C:野生型(WT)与突变体elm1在北京试验田的花期比较(长日照),***表示P<0.001显著差异;D:野生型(WT)与突变体elm1在海南三亚试验田的花期比较(短日照),ns表示无差异
Fig. 1 Phenotypic of the flowering period between WT and elm1A: Comparison of wild type (WT) and elm1 mutant at the flowering stage in the experimental field in Beijing (Bar=20 cm). B: Comparison of WT and elm1 mutant at the flowering stage in Sanya, Hainan (Bar=20 cm). C: Comparison of heading date between WT and elm1 mutant in the experimental field in Beijing (long-day), *** indicates a significant difference at P<0.001. D: Comparison of heading date between WT and elm1 mutant in the experimental field in Sanya, Hainan (short-day), ns stands for not significant
性状类别 Trait category | 旗叶叶长 Flag leaf length (cm) | 旗叶叶宽 Flag leaf width (cm) | 株高 Plant height (cm) | 分蘖数 Tiller number | 千粒重 Thousand-grain weight (g) | 每穗粒数 Grain number per panicle |
|---|---|---|---|---|---|---|
| WT | 28.15±1.76a | 1.56±0.13a | 84.36±1.23a | 14±2a | 24.86±0.58a | 110±3a |
| elm1 | 29.56±1.13a | 1.47±0.17a | 80.58±2.16a | 11±3a | 25.13±0.67a | 120±2a |
表1 野生型与elm1突变体的农艺性状比较(海南)
Table 1 Comparison of agronomic traits between WT and elm1 mutant (Hainan)
性状类别 Trait category | 旗叶叶长 Flag leaf length (cm) | 旗叶叶宽 Flag leaf width (cm) | 株高 Plant height (cm) | 分蘖数 Tiller number | 千粒重 Thousand-grain weight (g) | 每穗粒数 Grain number per panicle |
|---|---|---|---|---|---|---|
| WT | 28.15±1.76a | 1.56±0.13a | 84.36±1.23a | 14±2a | 24.86±0.58a | 110±3a |
| elm1 | 29.56±1.13a | 1.47±0.17a | 80.58±2.16a | 11±3a | 25.13±0.67a | 120±2a |
统计类别 Statistical category | 日本晴× elm1 Nipponbare×elm1 | elm1×日本晴 elm1×Nipponbare |
|---|---|---|
植株总数 Total number of plants | 396 | 374 |
正常植株数 Number of normal plants | 288 | 276 |
不开花植株数 Number of non-flowering plants | 108 | 98 |
分离比 Segregation ratio | 3∶1 | 1∶3 |
| χ2 | 0.066 7 | 0.051 2 |
表2 F2代分离群体统计结果
Table 2 Statistical results of F2 generation segregation population
统计类别 Statistical category | 日本晴× elm1 Nipponbare×elm1 | elm1×日本晴 elm1×Nipponbare |
|---|---|---|
植株总数 Total number of plants | 396 | 374 |
正常植株数 Number of normal plants | 288 | 276 |
不开花植株数 Number of non-flowering plants | 108 | 98 |
分离比 Segregation ratio | 3∶1 | 1∶3 |
| χ2 | 0.066 7 | 0.051 2 |
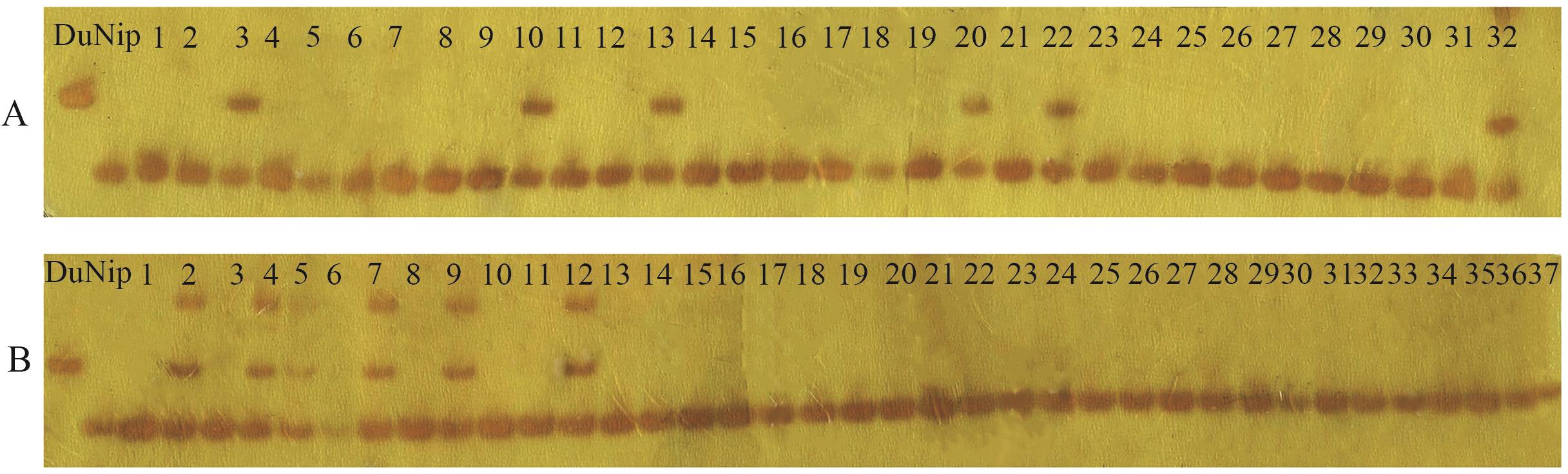
图2 突变体elm1基因连锁分析部分电泳图A:Indel-3标记在F2代单株的连锁分析(1‒32:32个F2代突变体表型单株的电泳条带结果);B:Indel-5标记在F2代单株的连锁分析(1‒37:37个F2代突变体表型单株的电泳条带结果);Nip:日本晴;Du:Dular
Fig. 2 Gel electrophoresis of gene linkage analysis for the mutant elm1A: Linkage analysis of the Indel-3 marker in individual F2 plants (1‒32: Electrophoresis banding results of 32 individual plants). B: Linkage analysis of the Indel-5 marker in individual F2 plants (1‒37: Electrophoresis banding results of 37 individual plants). Nip: Nipponbare. Du: Dular
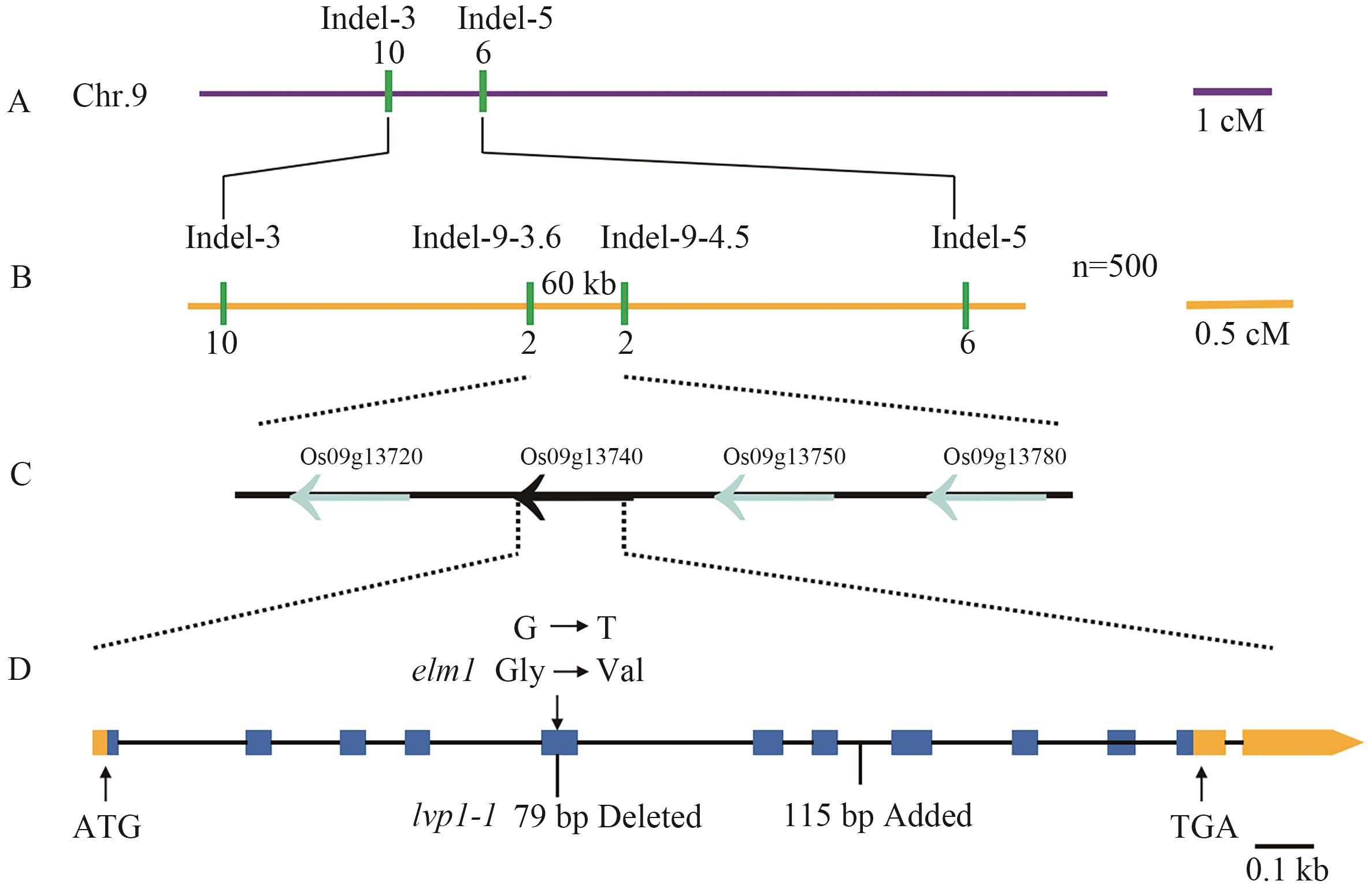
图3 基因ELM1图位克隆A:突变体elm1的基因初定位,每个标记之间的距离以重组频率(以cM为单位)表示;B:突变体elm1的基因精细定位;C:候选区域;D:突变体elm1突变发生LOC_Os09g13740基因的第5外显子,其中lvp1-1突变发生在第5外显子79 bp碱基的缺失,第8外显子与第9外显子之间插入115 bp碱基的突变
Fig. 3 Map-based cloning of gene ELM1A: Preliminary gene localization of elm1 mutant. The distance between each marker is represented by the recombination frequency, measured in centimorgans (cM). B: The fine mapping of the gene in the mutant elm1. C: Candidate region. D: The mutation in mutant elm1 occurs in the fifth exon of the LOC_Os09g13740, where the lvp1-1 mutation was a deletion of 79 base pairs in the fifth exon, and a mutation involving the insertion of 115 base pairs between the eighth and ninth exons

图4 野生型与突变体蛋白ELM1的结构预测A:野生型的ELM1蛋白结构预测;B:突变后ELM1的蛋白结构预测
Fig. 4 Prediction of the ELM1 protein structure in WT and mutantA: Prediction of the ELM1 protein structure in the wild type. B: Prediction of the ELM1 protein structure after mutation
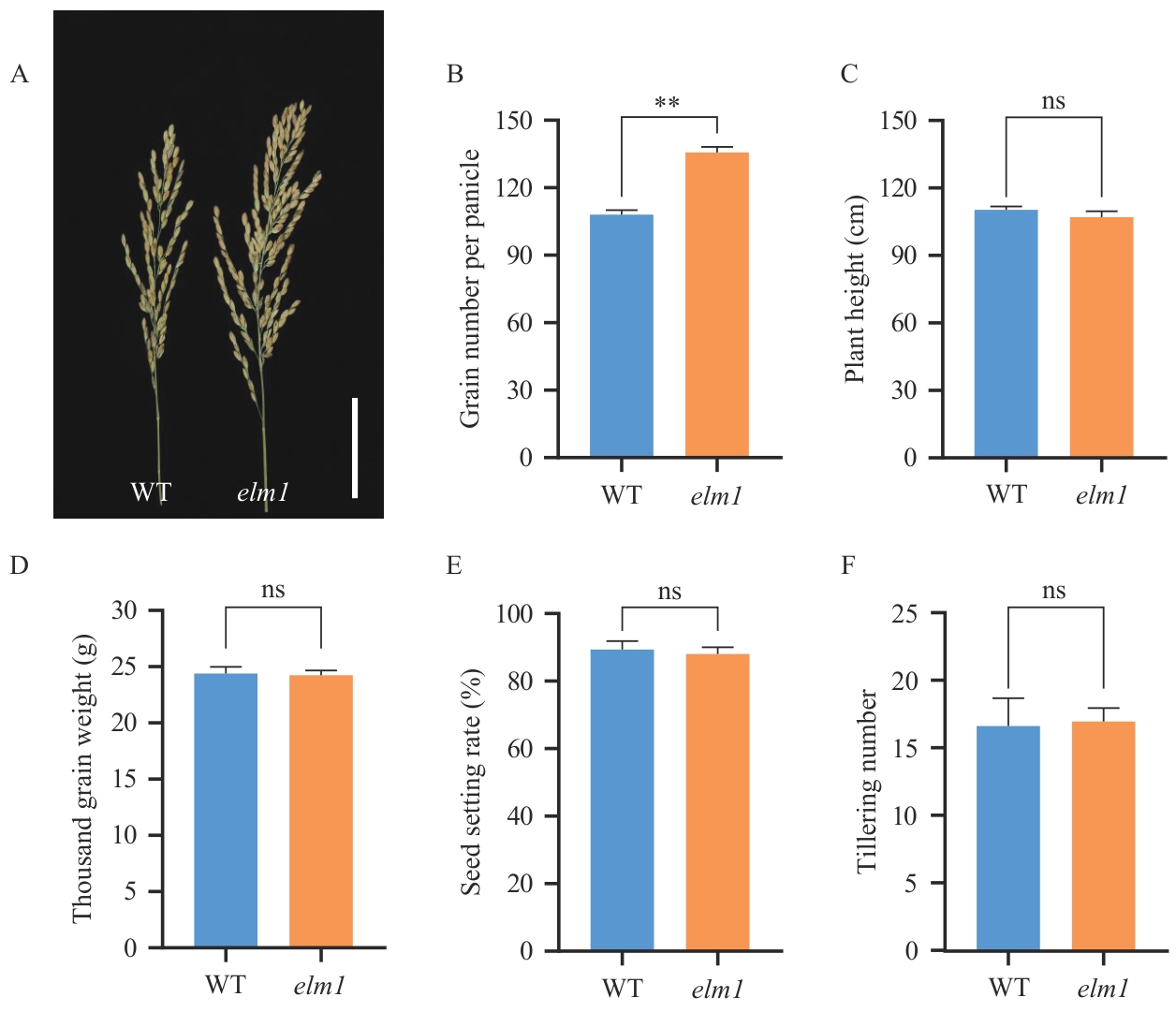
图5 野生型与突变体elm1的农艺性状比较A:野生型与elm1突变体的穗型比较(Bar=5 cm);B:野生型与elm1突变体的每穗粒数比较,**表示P<0.001显著差异;C:野生型与elm1突变体的株高比较;D:野生型与elm1突变体的千粒重比较;E:野生型与elm1突变体的结实率比较;F:野生型与elm1突变体的分蘖数比较
Fig. 5 Agronomic traits comparison between wild type and elm1 mutantA: Panicle comparison between WT and elm1 mutant (Bar=5 cm). B: Comparison of grain number per panicle between WT and elm1 mutant. ** indicates a significant difference at P<0.001. C: Comparison of plant height between WT and elm1 mutant. D: Comparison of thousand-grain weight between WT and elm1 mutant. E: Comparison of seed setting rate between WT and elm1 mutant. F: Comparison of tiller number between WT and elm1 mutant
| 1 | Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues [J]. Nat Rev Genet, 2012, 13(9): 627-639. |
| 2 | Song YH, Smith RW, To BJ, et al. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering [J]. Science, 2012, 336(6084): 1045-1049. |
| 3 | Wang JW. Regulation of flowering time by the miR156-mediated age pathway [J]. J Exp Bot, 2014, 65(17): 4723-4730. |
| 4 | Zhang SN, Zhang YY, Li KN, et al. Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice [J]. Curr Biol, 2021, 31(4): 671-683.e5. |
| 5 | Izawa T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice [J]. J Exp Bot, 2007, 58(12): 3091-3097. |
| 6 | Yano M, Katayose Y, Ashikari M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS [J]. Plant Cell, 2000, 12(12): 2473-2484. |
| 7 | Blümel M, Dally N, Jung C. Flowering time regulation in crops—what did we learn from Arabidopsis? [J]. Curr Opin Biotechnol, 2015, 32: 121-129. |
| 8 | Ga Z, Gao LY, Quzong XR, et al. Metabolomics, phytohormone and transcriptomics strategies to reveal the mechanism of barley heading date regulation to responds different photoperiod [J]. BMC Genomics, 2024, 25(1): 879. |
| 9 | Zhang ZY, Zhang B, Qi FX, et al. Hd1 function conversion in regulating heading is dependent on gene combinations of Ghd7, Ghd8, and Ghd7.1 under long-day conditions in rice [J]. Mol Breed, 2019, 39(7): 92. |
| 10 | Cui YX, Wang JR, Feng L, et al. A combination of long-day suppressor genes contributes to the northward expansion of rice [J]. Front Plant Sci, 2020, 11: 864. |
| 11 | Brambilla V, Fornara F. Y flowering? Regulation and activity of CONSTANS and CCT-domain proteins in Arabidopsis and crop species [J]. Biochim Biophys Acta Gene Regul Mech, 2017, 1860(5): 655-660. |
| 12 | Song YH, Shim JS, Kinmonth-Schultz HA, et al. Photoperiodic flowering: time measurement mechanisms in leaves [J]. Annu Rev Plant Biol, 2015, 66: 441-464. |
| 13 | Liu B, Liu YH, Wang BH, et al. The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment [J]. Nat Commun, 2019, 10(1): 2999. |
| 14 | Sun CH, Fang J, Zhao TL, et al. The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice [J]. Plant Cell, 2012, 24(8): 3235-3247. |
| 15 | Zong WB, Ren D, Huang MH, et al. Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1, Ghd7 and DTH8 in rice heading [J]. New Phytol, 2021, 229(3): 1635-1649. |
| 16 | Komiya R, Ikegami A, Tamaki S, et al. Hd3a and RFT1 are essential for flowering in rice [J]. Development, 2008, 135(4): 767-774. |
| 17 | Zheng QQ, Zhou ZJ, Li XR, et al. Heading date 3a stimulates tiller bud outgrowth in Oryza sativa L. through strigolactone signaling pathway [J]. Int J Mol Sci, 2024, 25(19): 10778. |
| 18 | Gao H, Zheng XM, Fei GL, et al. Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice [J]. PLoS Genet, 2013, 9(2): e1003281. |
| 19 | Zhang XN, Feng Q, Miao JS, et al. The WD40 domain-containing protein Ehd5 positively regulates flowering in rice (Oryza sativa) [J]. Plant Cell, 2023, 35(11): 4002-4019. |
| 20 | Wan SY, Wu JX, Zhang ZG, et al. Activation tagging, an efficient tool for functional analysis of the rice genome [J]. Plant Mol Biol, 2009, 69(1/2): 69-80. |
| 21 | Xue WY, Xing YZ, Weng XY, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice [J]. Nat Genet, 2008, 40(6): 761-767. |
| 22 | Ishimaru T, Hlaing KT, Oo YM, et al. An early-morning flowering trait in rice can enhance grain yield under heat stress field conditions at flowering stage [J]. Field Crops Res, 2022, 277: 108400. |
| 23 | Zhou SR, Zhu SS, Cui S, et al. Transcriptional and post-transcriptional regulation of heading date in rice [J]. New Phytol, 2021, 230(3): 943-956. |
| 24 | Yan WH, Wang P, Chen HX, et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice [J]. Mol Plant, 2011, 4(2): 319-330. |
| 25 | Wei XJ, Xu JF, Guo HN, et al. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously [J]. Plant Physiol, 2010, 153(4): 1747-1758. |
| 26 | Brambilla V, Gomez-Ariza J, Cerise M, et al. The importance of being on time: regulatory networks controlling photoperiodic flowering in cereals [J]. Front Plant Sci, 2017, 8: 665. |
| 27 | Jung C, Müller AE. Flowering time control and applications in plant breeding [J]. Trends Plant Sci, 2009, 14(10): 563-573. |
| 28 | Song J, Tang LQ, Cui YT, et al. Research progress on photoperiod gene regulation of heading date in rice [J]. Curr Issues Mol Biol, 2024, 46(9): 10299-10311. |
| 29 | Bai XF, Huang Y, Hu Y, et al. Duplication of an upstream silencer of FZP increases grain yield in rice [J]. Nat Plants, 2017, 3(11): 885-893. |
| 30 | Liu L, Gallagher J, Arevalo ED, et al. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes [J]. Nat Plants, 2021, 7(3): 287-294. |
| [1] | 陈晓军, 惠建, 马洪文, 白海波, 钟楠, 李稼润, 樊云芳. 利用单碱基基因编辑技术创制OsALS抗除草剂水稻种质资源[J]. 生物技术通报, 2025, 41(4): 106-114. |
| [2] | 李欣芃, 张武汉, 张莉, 舒服, 何强, 郭杨, 邓华凤, 王悦, 孙平勇. γ射线诱变创制水稻突变体及其分子鉴定[J]. 生物技术通报, 2025, 41(3): 35-43. |
| [3] | 方慧敏, 顾艺枢, 张晶, 张龙. 水稻叶片淀粉的分离与理化性质分析[J]. 生物技术通报, 2025, 41(2): 51-57. |
| [4] | 金素奎, 国倩倩, 刘巧泉, 高继平. 一种水稻叶片基因组DNA简易提取方法[J]. 生物技术通报, 2025, 41(1): 74-84. |
| [5] | 朱诗斐, 刘敬, 张家芊, 黄文坤, 彭德良, 孔令安, 彭焕. 水稻和拟禾本科根结线虫互作分子机制研究进展[J]. 生物技术通报, 2024, 40(9): 172-180. |
| [6] | 刘文志, 贺丹, 李鹏, 傅应林, 张译心, 温华杰, 于文清. 多粘类芽胞杆菌新菌株X-11及其对番茄和水稻的促生效应[J]. 生物技术通报, 2024, 40(9): 249-259. |
| [7] | 李庆懋, 彭聪归, 齐笑含, 刘兴蕾, 李臻园, 李沁妍, 黄立钰. 促进水稻铁素吸收的野生稻内生细菌优良菌株的筛选与鉴定[J]. 生物技术通报, 2024, 40(8): 255-263. |
| [8] | 孙志勇, 杜怀东, 刘阳, 马嘉欣, 于雪然, 马伟, 姚鑫杰, 王敏, 李培富. 水稻籽粒γ-氨基丁酸含量的全基因组关联分析[J]. 生物技术通报, 2024, 40(8): 53-62. |
| [9] | 庞梦真, 徐汉琴, 刘海燕, 宋娟, 王佳涵, 孙丽娜, 姬佩梅, 尹泽芝, 胡又川, 赵晓萌, 梁闪闪, 张泗举, 栾维江. 水稻黄化早抽穗突变体 hz1 的基因鉴定及功能分析[J]. 生物技术通报, 2024, 40(7): 125-136. |
| [10] | 田胜尼, 张琴, 董玉飞, 丁洲, 叶爱华, 张明珠. 酸性矿山废水对成熟期水稻根区理化因子及固氮微生物的影响[J]. 生物技术通报, 2024, 40(6): 271-280. |
| [11] | 杜兵帅, 邹昕蕙, 王子豪, 张馨元, 曹一博, 张凌云. 油茶SWEET基因家族的全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 179-190. |
| [12] | 彭凤, 余海霞, 张坤, 刘颖颖, 谭桂玉. 植物油体钙蛋白调控脂滴功能的研究进展[J]. 生物技术通报, 2024, 40(4): 33-39. |
| [13] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [14] | 李兴容, 谭志兵, 赵燕, 李曜魁, 赵炳然, 唐丽. 水稻低亲和性阳离子转运蛋白基因OsLCT3的克隆与功能研究[J]. 生物技术通报, 2024, 40(4): 97-109. |
| [15] | 李雪, 李容欧, 孔美懿, 黄磊. 解淀粉芽孢杆菌SQ-2对水稻的促生作用[J]. 生物技术通报, 2024, 40(2): 109-119. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||