








生物技术通报 ›› 2025, Vol. 41 ›› Issue (4): 188-197.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0964
• 研究报告 • 上一篇
收稿日期:2024-09-30
出版日期:2025-04-26
发布日期:2025-04-25
通讯作者:
于舒怡,男,博士,研究员,研究方向 :园艺作物病害综合防控;E-mail: crea0115@163.com作者简介:刘丽,女,博士,副研究员,研究方向 :园艺作物病害综合防控;E-mail: ll2006lx@163.com
基金资助:
LIU Li( ), WANG Hui, GUAN Tian-shu, LI Bai-hong, YU Shu-yi(
), WANG Hui, GUAN Tian-shu, LI Bai-hong, YU Shu-yi( )
)
Received:2024-09-30
Published:2025-04-26
Online:2025-04-25
摘要:
目的 ABA受体PYRl/PYLs/RCARs在ABA信号转导通路中起着重要作用。通过酵母双杂交技术筛选葡萄VvPYL4的互作蛋白,探究VvPYL4在葡萄应答霜霉病菌胁迫信号通路中的作用。 方法 以霜霉病菌侵染‘贝达’葡萄叶片为材料,构建cDNA文库;构建诱饵表达载体pGBKT7-VvPYL4,通过酵母双杂交技术从cDNA文库中筛选与VvPYL4相互作用的蛋白;通过实时荧光定量PCR分析候选互作蛋白基因在葡萄霜霉病菌诱导下的表达模式,并通过双分子荧光互补技术进行互作蛋白的验证。 结果 构建的酵母双杂交cDNA文库库容为7.16×107 CFU/mL,重组率100%,插入片段大小在1 000 bp左右。成功构建诱饵表达载体pGBKT7-VvPYL4,且在酵母细胞中无自激活活性。诱饵载体与酵母双杂交文库共转酵母AH109菌株后,经多次筛库、测序、BLAST比对和回转验证,最终获得53个候选互作蛋白,这些蛋白涉及信号转导、植物生长发育及环境胁迫响应等多个方面。基于实时荧光定量PCR分析,编码4个蛋白的基因均受葡萄霜霉病菌诱导表达。通过双分子荧光互补试验发现,在共转pSPYCE-PP2C24和pSPYNE-PYL4表达载体的本氏烟草叶片中可观察到强烈的黄色荧光信号,表明PYL4与PP2C24蛋白之间能够发生相互作用。 结论 成功构建霜霉病菌侵染葡萄叶片的cDNA文库,并筛选出53个与VvPYL4相互作用的候选蛋白,其中4个编码蛋白的基因均响应葡萄霜霉病菌的胁迫诱导,验证了VvPYL4与PP2C24蛋白之间存在相互作用关系。
刘丽, 王辉, 关天舒, 李柏宏, 于舒怡. 葡萄脱落酸受体VvPYL4互作蛋白的筛选及互作蛋白基因表达[J]. 生物技术通报, 2025, 41(4): 188-197.
LIU Li, WANG Hui, GUAN Tian-shu, LI Bai-hong, YU Shu-yi. Screening the Interacting Protein of Abscisic Acid Receptor VvPYL4 and the Gene Expression of the Interacting Protein in Grape[J]. Biotechnology Bulletin, 2025, 41(4): 188-197.
| 基因 Gene | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) |
|---|---|---|
| VvMYB3R-3 | CTTTCCCAATACTCCTTCCA | CCCAGTTCTTTCTTGTTCATC |
| VvPP2C24 | CTCGTGTTCCTCTTTCGTTC | CATTCTTCCAACCTGACTCC |
| VvASR1 | CCTACTCCGACACCACCTAC | GTGCTTCTCCTCCTTCCTG |
| VvPOD6 | TGCAGTGAAACTTCTTCAGC | CATCCACCCTTCCATACTTC |
| EF1a | GAACTGGGTGCTTGATAGGC | ACCAAAATATCCGGAGTAAAAGA |
表1 RT-qPCR引物
Table 1 Primers used in RT-qPCR
| 基因 Gene | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) |
|---|---|---|
| VvMYB3R-3 | CTTTCCCAATACTCCTTCCA | CCCAGTTCTTTCTTGTTCATC |
| VvPP2C24 | CTCGTGTTCCTCTTTCGTTC | CATTCTTCCAACCTGACTCC |
| VvASR1 | CCTACTCCGACACCACCTAC | GTGCTTCTCCTCCTTCCTG |
| VvPOD6 | TGCAGTGAAACTTCTTCAGC | CATCCACCCTTCCATACTTC |
| EF1a | GAACTGGGTGCTTGATAGGC | ACCAAAATATCCGGAGTAAAAGA |

图4 VvPYL4互作蛋白筛选+为阳性对照:pGBKT7-p53+pGADT7-largeT
Fig. 4 Screening of VvPYL4 interacting proteins"+" indicates the positive control: pGBKT7-p53+pGADT7-largeT
| 编号 No. | 酵母克隆编号 Code of yeast clone | 基因编号 Gene ID | 功能注释 Function annotation | 筛选次数 Number of screenings |
|---|---|---|---|---|
| 1 | BD-VvPYL4-4 | LOC117926668 | Thaumatin-like protein 1 | 1 |
| 2 | BD-VvPYL4-6 | LOC100245034 | Hypersensitive-induced reaction 1 protein | 1 |
| 3 | BD-VvPYL4-10 | LOC117928201 | Small heat shock protein | 1 |
| 4 | BD-VvPYL4-12 | LOC100256181 | Transcription factor MYB3R-3 | 1 |
| 5 | BD-VvPYL4-13 | LOC100260476 | Pathogenesis-related protein PR-4 | 2 |
| 6 | BD-VvPYL4-16 | LOC117913055 | Glutamate-glyoxylate aminotransferase 2 | 1 |
| 7 | BD-VvPYL4-18 | LOC100246378 | Glyceraldehyde-3-phosphate dehydrogenase A | 1 |
| 8 | BD-VvPYL4-19 | LOC100248362 | Mitochondrial outer membrane protein porin of 36 kDa | 1 |
| 9 | BD-VvPYL4-20 | LOC100257132 | Glycine rich protein family | 1 |
| 10 | BD-VvPYL4-21 | LOC100245393 | Chlorophyll A-B binding protein | 1 |
| 11 | BD-VvPYL4-22 | LOC100268135 | Chaperone protein dnaJ A6 | 1 |
| 12 | BD-VvPYL4-23 | LOC100249605 | Expansin-A13 | 1 |
| 13 | BD-VvPYL4-25 | LOC117905970 | Thioredoxin-like protein CDSP32 | 1 |
| 14 | BD-VvPYL4-26 | LOC100255251 | Protein phosphatase 2C 24 | 2 |
| 15 | BD-VvPYL4-27 | LOC100258879 | Cytochrome b6-f complex iron-sulfur subunit 1 | 3 |
| 16 | BD-VvPYL4-29 | LOC100233124 | RD22-like protein | 3 |
| 17 | BD-VvPYL4-30 | LOC100254430 | Protein DEHYDRATION-INDUCED 19 homolog 3 | 2 |
| 18 | BD-VvPYL4-31 | LOC100251062 | ATP synthase delta chain | 1 |
| 19 | BD-VvPYL4-33 | LOC117916106 | Ribulose bisphosphate carboxylase/oxygenase activase | 2 |
| 20 | BD-VvPYL4-34 | LOC117919543 | Probable galactinol-sucrose galactosyltransferase 2 | 1 |
| 21 | BD-VvPYL4-35 | LOC100526774 | Cytochrome P450 | 1 |
| 22 | BD-VvPYL4-36 | LOC100852851 | Fructose-bisphosphate aldolase 1 | 1 |
| 23 | BD-VvPYL4-37 | LOC100247997 | Anthocyanidin 3-O-glucosyltransferase 5 | 1 |
| 24 | BD-VvPYL4-38 | LOC117932928 | Ethylene-responsive transcription factor 5-like | 1 |
| 25 | BD-VvPYL4-39 | LOC117906248 | Purine permease 1-like | 1 |
| 26 | BD-VvPYL4-40 | LOC100254009 | Peroxidase 6 | 1 |
| 27 | BD-VvPYL4-41 | LOC100240756 | Endo-1, 3; 1, 4-beta-D-glucanase-like | 1 |
| 28 | BD-VvPYL4-42 | LOC100241982 | Phospholipase D delta | 1 |
| 29 | BD-VvPYL4-44 | LOC100264642 | Probable 3-hydroxyisobutyrate dehydrogenase-like 1 | 1 |
| 30 | BD-VvPYL4-45 | LOC100245816 | Methyl-CpG-binding domain-containing protein 9 | 1 |
| 31 | BD-VvPYL4-47 | LOC100255748 | Uncharacterized | 1 |
| 32 | BD-VvPYL4-50 | LOC100267253 | Metallothionein | 1 |
| 33 | BD-VvPYL4-52 | LOC100260811 | Oxygen-evolving enhancer protein 1 | 1 |
| 34 | BD-VvPYL4-53 | LOC100253703 | Rac-like GTP-binding protein RAC1 | 1 |
| 35 | BD-VvPYL4-54 | LOC100250281 | Abscisic stress-ripening protein 1-like | 1 |
| 36 | BD-VvPYL4-55 | LOC117911472 | Ferredoxin--nitrite reductase | 1 |
| 37 | BD-VvPYL4-58 | LOC100853165 | Catalase-related immune-responsive | 1 |
| 38 | BD-VvPYL4-60 | LOC100267892 | 20 kD chaperonin | 1 |
| 39 | BD-VvPYL4-61 | LOC117905125 | Protein P21-like | 4 |
| 40 | BD-VvPYL4-62 | LOC100247939 | Cinnamoyl-CoA reductase 1 | 1 |
| 41 | BD-VvPYL4-64 | LOC100252685 | Leucine Rich repeat | 1 |
| 42 | BD-VvPYL4-65 | LOC100264042 | Tim17/Tim22/Tim23/Pmp24 family | 1 |
| 43 | BD-VvPYL4-69 | LOC100249137 | Beta carbonic anhydrase 1 | 1 |
| 44 | BD-VvPYL4-74 | LOC117928074 | Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit A-like | 1 |
| 45 | BD-VvPYL4-76 | LOC100855103 | Domain of unknown function | 1 |
| 46 | BD-VvPYL4-77 | LOC100245544 | Bifunctional monothiol glutaredoxin-S16 | 1 |
| 47 | BD-VvPYL4-78 | LOC117907066 | Elongation factor 1-gamma | 1 |
| 48 | BD-VvPYL4-79 | LOC100264196 | Cysteine synthase | 3 |
| 49 | BD-VvPYL4-83 | LOC117915980 | Elongation factor 1-alpha | 1 |
| 50 | BD-VvPYL4-85 | LOC100267386 | Aldehyde dehydrogenase family | 1 |
| 51 | BD-VvPYL4-86 | LOC117904623 | Chaperone protein ClpB1 | 1 |
| 52 | BD-VvPYL4-87 | LOC117924474 | Probable galactinol-sucrose galactosyltransferase 6 | 1 |
| 53 | BD-VvPYL4-88 | LOC100243967 | Photosystem II 10 kD polypeptide PsbR | 1 |
表2 VvPYL4互作蛋白的注释结果
Table 2 Annotation results of VvPYL4 interaction protein
| 编号 No. | 酵母克隆编号 Code of yeast clone | 基因编号 Gene ID | 功能注释 Function annotation | 筛选次数 Number of screenings |
|---|---|---|---|---|
| 1 | BD-VvPYL4-4 | LOC117926668 | Thaumatin-like protein 1 | 1 |
| 2 | BD-VvPYL4-6 | LOC100245034 | Hypersensitive-induced reaction 1 protein | 1 |
| 3 | BD-VvPYL4-10 | LOC117928201 | Small heat shock protein | 1 |
| 4 | BD-VvPYL4-12 | LOC100256181 | Transcription factor MYB3R-3 | 1 |
| 5 | BD-VvPYL4-13 | LOC100260476 | Pathogenesis-related protein PR-4 | 2 |
| 6 | BD-VvPYL4-16 | LOC117913055 | Glutamate-glyoxylate aminotransferase 2 | 1 |
| 7 | BD-VvPYL4-18 | LOC100246378 | Glyceraldehyde-3-phosphate dehydrogenase A | 1 |
| 8 | BD-VvPYL4-19 | LOC100248362 | Mitochondrial outer membrane protein porin of 36 kDa | 1 |
| 9 | BD-VvPYL4-20 | LOC100257132 | Glycine rich protein family | 1 |
| 10 | BD-VvPYL4-21 | LOC100245393 | Chlorophyll A-B binding protein | 1 |
| 11 | BD-VvPYL4-22 | LOC100268135 | Chaperone protein dnaJ A6 | 1 |
| 12 | BD-VvPYL4-23 | LOC100249605 | Expansin-A13 | 1 |
| 13 | BD-VvPYL4-25 | LOC117905970 | Thioredoxin-like protein CDSP32 | 1 |
| 14 | BD-VvPYL4-26 | LOC100255251 | Protein phosphatase 2C 24 | 2 |
| 15 | BD-VvPYL4-27 | LOC100258879 | Cytochrome b6-f complex iron-sulfur subunit 1 | 3 |
| 16 | BD-VvPYL4-29 | LOC100233124 | RD22-like protein | 3 |
| 17 | BD-VvPYL4-30 | LOC100254430 | Protein DEHYDRATION-INDUCED 19 homolog 3 | 2 |
| 18 | BD-VvPYL4-31 | LOC100251062 | ATP synthase delta chain | 1 |
| 19 | BD-VvPYL4-33 | LOC117916106 | Ribulose bisphosphate carboxylase/oxygenase activase | 2 |
| 20 | BD-VvPYL4-34 | LOC117919543 | Probable galactinol-sucrose galactosyltransferase 2 | 1 |
| 21 | BD-VvPYL4-35 | LOC100526774 | Cytochrome P450 | 1 |
| 22 | BD-VvPYL4-36 | LOC100852851 | Fructose-bisphosphate aldolase 1 | 1 |
| 23 | BD-VvPYL4-37 | LOC100247997 | Anthocyanidin 3-O-glucosyltransferase 5 | 1 |
| 24 | BD-VvPYL4-38 | LOC117932928 | Ethylene-responsive transcription factor 5-like | 1 |
| 25 | BD-VvPYL4-39 | LOC117906248 | Purine permease 1-like | 1 |
| 26 | BD-VvPYL4-40 | LOC100254009 | Peroxidase 6 | 1 |
| 27 | BD-VvPYL4-41 | LOC100240756 | Endo-1, 3; 1, 4-beta-D-glucanase-like | 1 |
| 28 | BD-VvPYL4-42 | LOC100241982 | Phospholipase D delta | 1 |
| 29 | BD-VvPYL4-44 | LOC100264642 | Probable 3-hydroxyisobutyrate dehydrogenase-like 1 | 1 |
| 30 | BD-VvPYL4-45 | LOC100245816 | Methyl-CpG-binding domain-containing protein 9 | 1 |
| 31 | BD-VvPYL4-47 | LOC100255748 | Uncharacterized | 1 |
| 32 | BD-VvPYL4-50 | LOC100267253 | Metallothionein | 1 |
| 33 | BD-VvPYL4-52 | LOC100260811 | Oxygen-evolving enhancer protein 1 | 1 |
| 34 | BD-VvPYL4-53 | LOC100253703 | Rac-like GTP-binding protein RAC1 | 1 |
| 35 | BD-VvPYL4-54 | LOC100250281 | Abscisic stress-ripening protein 1-like | 1 |
| 36 | BD-VvPYL4-55 | LOC117911472 | Ferredoxin--nitrite reductase | 1 |
| 37 | BD-VvPYL4-58 | LOC100853165 | Catalase-related immune-responsive | 1 |
| 38 | BD-VvPYL4-60 | LOC100267892 | 20 kD chaperonin | 1 |
| 39 | BD-VvPYL4-61 | LOC117905125 | Protein P21-like | 4 |
| 40 | BD-VvPYL4-62 | LOC100247939 | Cinnamoyl-CoA reductase 1 | 1 |
| 41 | BD-VvPYL4-64 | LOC100252685 | Leucine Rich repeat | 1 |
| 42 | BD-VvPYL4-65 | LOC100264042 | Tim17/Tim22/Tim23/Pmp24 family | 1 |
| 43 | BD-VvPYL4-69 | LOC100249137 | Beta carbonic anhydrase 1 | 1 |
| 44 | BD-VvPYL4-74 | LOC117928074 | Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit A-like | 1 |
| 45 | BD-VvPYL4-76 | LOC100855103 | Domain of unknown function | 1 |
| 46 | BD-VvPYL4-77 | LOC100245544 | Bifunctional monothiol glutaredoxin-S16 | 1 |
| 47 | BD-VvPYL4-78 | LOC117907066 | Elongation factor 1-gamma | 1 |
| 48 | BD-VvPYL4-79 | LOC100264196 | Cysteine synthase | 3 |
| 49 | BD-VvPYL4-83 | LOC117915980 | Elongation factor 1-alpha | 1 |
| 50 | BD-VvPYL4-85 | LOC100267386 | Aldehyde dehydrogenase family | 1 |
| 51 | BD-VvPYL4-86 | LOC117904623 | Chaperone protein ClpB1 | 1 |
| 52 | BD-VvPYL4-87 | LOC117924474 | Probable galactinol-sucrose galactosyltransferase 6 | 1 |
| 53 | BD-VvPYL4-88 | LOC100243967 | Photosystem II 10 kD polypeptide PsbR | 1 |
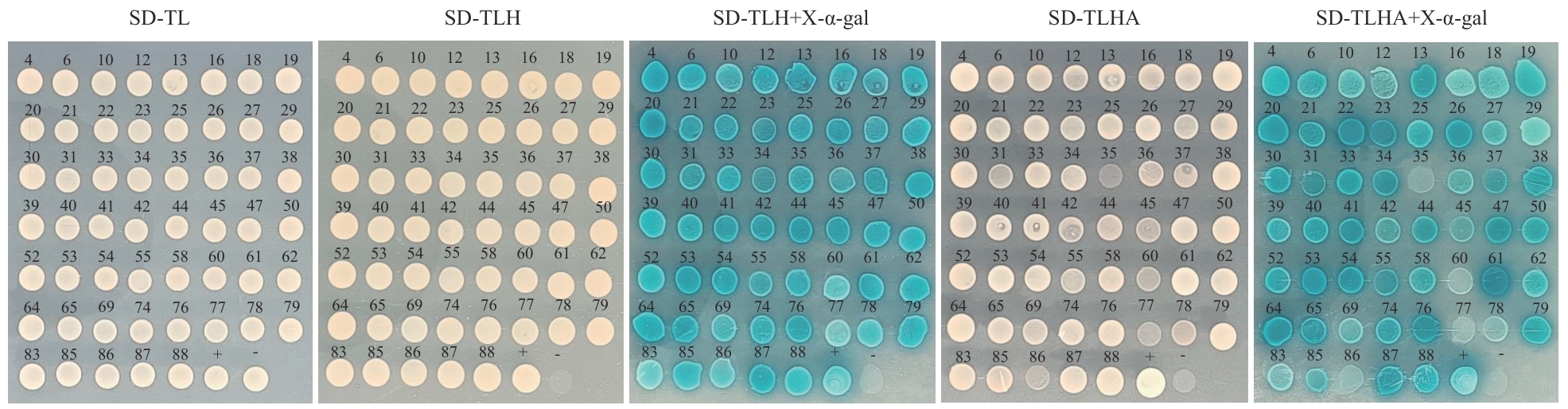
图5 回转验证结果+为阳性对照pGBKT7-p53+pGADT7-largeT;-为阴性对照pGBKT7-laminC+pGADT7-largeT
Fig. 5 Results of rotation verification"+" indicates the positive control pGBKT7-p53+pGADT7-largeT; "-" indicates the negative control pGBKT7-laminC+pGADT7-largeT
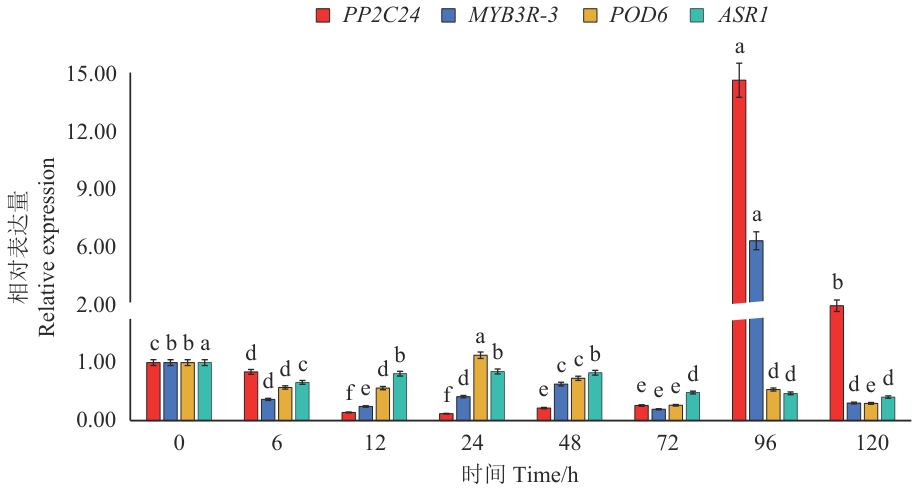
图6 霜霉病菌处理下VvPYL4互作蛋白基因的表达分析不同小写字母表示在0.05水平上具有显著性差异
Fig. 6 Expressions of the VvPYL4-interacting protein genes under Plasmopara viticola stressDifferent lowercase letters indicate significant differences at the 0.05 level
| 29 | Gao XH, Jia RY, Wang MS, et al. Construction and identification of a cDNA library for use in the yeast two-hybrid system from duck embryonic fibroblast cells post-infected with duck enteritis virus [J]. Mol Biol Rep, 2014, 41(1): 467-475. |
| 30 | 刘露露, 曲俊杰, 郭泽西, 等. 霜霉菌侵染后葡萄叶片酵母双杂交cDNA文库构建 [J]. 南方农业学报, 2020, 51(4): 829-835. |
| Liu LL, Qu JJ, Guo ZX, et al. Construction of a yeast two-hybrid cDNA library from Vitis vinifera leaves infected by downy mildew [J]. J South Agric, 2020, 51(4): 829-835. | |
| 31 | Das PP, Macharia MW, Lin QS, et al. In planta proximity-dependent biotin identification (BioID) identifies a TMV replication co-chaperone NbSGT1 in the vicinity of 126 kDa replicase [J]. J Proteomics, 2019, 204: 103402. |
| 32 | Mei Q, Li S, Liu P, et al. Construction and identification of a yeast two-hybrid bait vector and its effect on the growth of yeast cells and the self-activating function of reporter genes for screening of HPV18 E6-interacting protein [J]. J Huazhong Univ Sci Technolog Med Sci, 2010, 30(1): 8-12. |
| 33 | Braun EL, Grotewold E. Newly discovered plant c-myb-like genes rewrite the evolution of the plant myb gene family [J]. Plant Physiol, 1999, 121(1): 21-24. |
| 34 | Wilkins O, Nahal H, Foong J, et al. Expansion and diversification of the Populus R2R3-MYB family of transcription factors [J]. Plant Physiol, 2009, 149(2): 981-993. |
| 35 | Wong DCJ, Schlechter R, Vannozzi A, et al. A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation [J]. DNA Res, 2016, 23(5): 451-466. |
| 36 | Huang S, Wang CL, Ding ZX, et al. A plant NLR receptor employs ABA central regulator PP2C-SnRK2 to activate antiviral immunity [J]. Nat Commun, 2024, 15(1): 3205. |
| 37 | 蒋选利, 李振岐, 康振生. 过氧化物酶与植物抗病性研究进展 [J]. 西北农林科技大学学报: 自然科学版, 2001, 29(6): 124-129. |
| Jiang XL, Li ZQ, Kang ZS. The recent progress of research on peroxidase in plant disease resistance [J]. J Northwest A F Univ Nat Sci Ed, 2001, 29(6): 124-129. | |
| 38 | 程维舜, 孙玉宏, 曾红霞, 等. ASR蛋白与植物的抗逆性研究进展 [J]. 园艺学报, 2013, 40(10): 2049-2057. |
| 1 | 张玮, 燕继晔, 刘梅, 等. 葡萄霜霉病流行与预测研究进展 [J]. 中国果树, 2020(3): 11-15. |
| Zhang W, Yan JY, Liu M, et al. Research progress on prediction and epidemic of grapevine downy mildew [J]. China Fruits, 2020(3): 11-15. | |
| 2 | Fields S, Song O. A novel genetic system to detect protein-protein interactions [J]. Nature, 1989, 340(6230): 245-246. |
| 3 | 陈玉婷, 刘露, 楚盼盼, 等. 受青枯菌诱导的花生根酵母双杂交文库构建和AhRRS5互作蛋白的筛选 [J]. 作物学报, 2021, 47(11): 2134-2146. |
| Chen YT, Liu L, Chu PP, et al. Construction of yeast two-hybrid cDNA library induced by Ralstonia solanacearum and interaction protein screening for AhRRS5 in peanut [J]. Acta Agron Sin, 2021, 47(11): 2134-2146. | |
| 4 | 张恒, 陈怡名, 张旭, 等. 白粉菌诱导簇毛麦叶片酵母双杂交文库构建及CMPG1-Ⅴ候选互作蛋白筛选 [J]. 南京农业大学学报, 2020, 43(4): 594-604. |
| Zhang H, Chen YM, Zhang X, et al. Construction of yeast two-hybrid cDNA library of Haynaldia villosa leaves induced by Blumeria graminis f. sp. tritici and candidate interaction protein screening for CMPG1-Ⅴ [J]. J Nanjing Agric Univ, 2020, 43(4): 594-604. | |
| 5 | Martin GB, Brommonschenkel SH, Chunwongse J, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato [J]. Science, 1993, 262(5138): 1432-1436. |
| 6 | Scofield SR, Tobias CM, Rathjen JP, et al. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato [J]. Science, 1996, 274(5295): 2063-2065. |
| 7 | 许向阳, 裴童, 吴泰茹, 等. Cf-19介导的抗番茄叶霉病(Cladosporium fulvum)免疫应答酵母双杂交cDNA文库构建和鉴定 [J]. 东北农业大学学报, 2020, 51(5): 10-16. |
| Xu XY, Pei T, Wu TR, et al. Construction and identification of yeast two-hybrid cDNA library for Cf-19 mediated resistance to Cladosporium fulvum infection in tomato [J]. J Northeast Agric Univ, 2020, 51(5): 10-16. | |
| 8 | MacKey D, Belkhadir Y, Alonso JM, et al. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance [J]. Cell, 2003, 112(3): 379-389. |
| 9 | 王玉姣, 陈姗姗, 孙柏华, 等. 辣椒疫霉菌诱导辣椒酵母双杂交cDNA文库的构建及鉴定 [J]. 山东农业大学学报: 自然科学版, 2018, 49(3): 379-382. |
| Wang YJ, Chen SS, Sun BH, et al. Construction and characterization of yeast two-hybrid cDNA library derived from Capsicum annuum inoculated with Phytophthora capsici [J]. J Shandong Agric Univ Nat Sci Ed, 2018, 49(3): 379-382. | |
| 10 | 郭文雅, 赵京献, 郭伟珍. 脱落酸(ABA)生物学作用研究进展 [J]. 中国农学通报, 2014, 30(21): 205-210. |
| Guo WY, Zhao JX, Guo WZ. Advance of research on biological function of abscisic acid (ABA) [J]. Chin Agric Sci Bull, 2014, 30(21): 205-210. | |
| 11 | 王超霞. 葡萄霜霉病程中白藜芦醇及其衍生物的积累及抗病机制 [D]. 北京: 中国农业大学, 2017. |
| Wang CX. Accumulation and resistant mechanism of resveratrol and derivatives during grape defense to downy mildew[D]. Beijing: China Agricultural University, 2017. | |
| 12 | Lackman P, González-Guzmán M, Tilleman S, et al. Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco [J]. Proc Natl Acad Sci U S A, 2011, 108(14): 5891-5896. |
| 13 | Li Y, Zhang XC, Jiang JB, et al. Virus-induced gene silencing of SlPYL4 decreases the drought tolerance of tomato [J]. Hortic Plant J, 2022, 8(3): 361-368. |
| 14 | Wang CC, Gao HJ, Chu ZH, et al. A nonspecific lipid transfer protein, StLTP10, mediates resistance to Phytophthora infestans in potato [J]. Mol Plant Pathol, 2021, 22(1): 48-63. |
| 15 | Cao JM, Jiang M, Li P, et al. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon [J]. BMC Genomics, 2016, 17: 175. |
| 16 | 杨冬冬. 藜麦PYL-PP2C-SnRK2s基因家族的鉴定及功能分析 [D]. 烟台: 烟台大学, 2024. |
| Yang DD. Identification and functional analysis of PYL-PP2C-SnRK2s gene family in quinoa (Chenopodium quinoa Willd.) [D]. Yantai: Yantai University, 2024. | |
| 17 | Xue TT, Wang D, Zhang SZ, et al. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis [J]. BMC Genomics, 2008, 9: 550. |
| 18 | 何红红, 路志浩, 马宗桓, 等. 葡萄PP2C家族基因的鉴定与表达分析 [J]. 园艺学报, 2018, 45(7): 1237-1250. |
| He HH, Lu ZH, Ma ZH, et al. Genome-wide identification and expression analysis of the PP2C gene family in Vitis vinifera [J]. Acta Hortic Sin, 2018, 45(7): 1237-1250. | |
| 19 | Liu L, Liu CY, Wang H, et al. The abscisic acid receptor gene VvPYL4 positively regulates grapevine resistance to Plasmopara viticola [J]. Plant Cell Tissue Organ Cult PCTOC, 2020, 142(3): 483-492. |
| 20 | Liu L, Zhang B, Wang H, et al. Candidate resistance genes selection and transcriptome analysis for the early responses to Plasmopara viticola infection in grape cultivars [J]. J Plant Pathol, 2020, 102(3): 857-869. |
| 21 | 刘丽, 李柏宏, 郑丽娇, 等. 葡萄脱落酸受体基因VvPYL4的生物信息学及表达分析 [J]. 沈阳农业大学学报, 2022, 53(3): 286-293. |
| Liu L, Li BH, Zheng LJ, et al. The bioinformatics and expression analysis of an abscisic acid receptor gene VvPYL4 in grapevine [J]. J Shenyang Agric Univ, 2022, 53(3): 286-293. | |
| 22 | 黄丽萍, 刘力宁, 蒋明义. 水稻锌指蛋白ZFP36互作蛋白的筛选及互作蛋白基因表达 [J]. 基因组学与应用生物学, 2022, 41(6): 1316-1328. |
| Huang LP, Liu LN, Jiang MY. Screening the interacting protein of ZFP36 and the expression of the interacting protein in Oryza sativa [J]. Genom Appl Biol, 2022, 41(6): 1316-1328. | |
| 23 | Miller JP, Lo RS, Ben-Hur A, et al. Large-scale identification of yeast integral membrane protein interactions [J]. Proc Natl Acad Sci U S A, 2005, 102(34): 12123-12128. |
| 24 | 王川, 盛鸥, 窦同心, 等. 香蕉果实酵母双杂交文库的构建及后熟转录因子MaMADS2互作蛋白筛选 [J]. 南京农业大学学报, 2023, 46(2): 237-246. |
| Wang C, Sheng O, Dou TX, et al. Construction of yeast two-hybrid library and screening of interacting proteins of ripening regulator MaMADS2 in banana fruit [J]. J Nanjing Agric Univ, 2023, 46(2): 237-246. | |
| 25 | He Y, Wu L, Liu X, et al. Yeast two-hybrid screening for proteins that interact with PFT in wheat [J]. Sci Rep, 2019, 9(1): 15521. |
| 26 | Song PW, Zhang LF, Wu LL, et al. A ricin B-like lectin protein physically interacts with TaPFT and is involved in resistance to Fusarium head blight in wheat [J]. Phytopathology, 2021, 111(12): 2309-2316. |
| 27 | 宾羽, 张琦, 王春庆, 等. 利用酵母双杂交系统筛选与柑橘黄化脉明病毒CP互作的寄主因子 [J]. 中国农业科学, 2023, 56(10): 1881-1892. |
| Bin Y, Zhang Q, Wang CQ, et al. Screening of the host factors interacting with CP of citrus yellow vein clearing virus by yeast two-hybrid system [J]. Sci Agric Sin, 2023, 56(10): 1881-1892. | |
| 28 | 马芳芳, 蒋明义. 外源ABA处理的玉米叶片酵母双杂交cDNA文库的构建及评价 [J]. 江苏农业科学, 2016, 44(4): 58-61. |
| Ma FF, Jiang MY. Construction and evaluation of yeast two hybrid cDNA library of maize leaf treated with exogenous ABA [J]. Jiangsu Agric Sci, 2016, 44(4): 58-61. | |
| 38 | Cheng WS, Sun YH, Zeng HX, et al. ASR protein and plant stress tolerance [J]. Acta Hortic Sin, 2013, 40(10): 2049-2057. |
| 39 | 陈雪琴, 庞倩倩, 裴丹, 等. 葡萄VvGCN5基因的鉴定及互作蛋白筛选 [J]. 南京农业大学学报, 2023, 46(5): 852-861. |
| Chen XQ, Pang QQ, Pei D, et al. Identification of grape VvGCN5 gene and screening of interacting proteins [J]. J Nanjing Agric Univ, 2023, 46(5): 852-861. | |
| 40 | 程梅. 玉米脱落酸受体(PYL3)与2C型丝氨/苏氨酸蛋白磷酸酶(PP2C)的互作分析 [D]. 雅安: 四川农业大学, 2014. |
| Cheng M. Interaction between abscisic acid receptor (PYL3) and serine/threonine protein phosphatase type 2C in maize[D]. Ya'an: Sichuan Agricultural University, 2014. | |
| 41 | Zhao Y, Xing L, Wang XG, et al. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes [J]. Sci Signal, 2014, 7(328): ra53. |
| 42 | Diao ZH, Yang RQ, Wang YZ, et al. Functional screening of the Arabidopsis 2C protein phosphatases family identifies PP2C15 as a negative regulator of plant immunity by targeting BRI1-associated receptor kinase 1 [J]. Mol Plant Pathol, 2024, 25(4): e13447. |
| [1] | 沈川, 李夏, 覃剑锋, 段龙飞, 刘佳. 基于软腐病菌诱导的魔芋酵母双杂交文库筛选WRKY72互作蛋白[J]. 生物技术通报, 2025, 41(1): 85-94. |
| [2] | 邢丽南, 张艳芳, 葛明然, 赵令敏, 陈妍, 霍秀文. 山药DoWRKY40基因表达特征分析及互作蛋白筛选[J]. 生物技术通报, 2024, 40(8): 118-128. |
| [3] | 王秋月, 段鹏亮, 李海笑, 刘宁, 曹志艳, 董金皋. 玉米大斑病菌cDNA文库的构建及转录因子StMR1互作蛋白的筛选[J]. 生物技术通报, 2024, 40(6): 281-289. |
| [4] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [5] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [6] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [7] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [8] | 蔡佳, 梁振宇, 黄瑜, 鲁义善, 施钢, 简纪常. 利用酵母双杂交系统筛选与鉴定石斑鱼EcBAG3互作因子[J]. 生物技术通报, 2022, 38(8): 77-83. |
| [9] | 刘娟, 朱春晓, 肖雪琼, 莫陈汨, 王高峰, 肖炎农. 淡紫紫孢菌亲环蛋白PlCYP6 互作蛋白的筛选[J]. 生物技术通报, 2021, 37(7): 137-145. |
| [10] | 杨华杰, 周玉萍, 范甜, 吕天晓, 谢楚萍, 田长恩. 拟南芥IQM4互作蛋白的筛选和鉴定[J]. 生物技术通报, 2021, 37(11): 190-196. |
| [11] | 郝小花, 戴佳利, 暨文劲, 黄丹, 李东屏, 田连福. 水稻籽粒低镉蛋白LCD互作蛋白的筛选与鉴定[J]. 生物技术通报, 2020, 36(11): 21-29. |
| [12] | 钟李婷, 陈秀珍, 唐云, 李俊仁, 王小兵, 刘彦婷, 周璇璇, 詹若挺, 陈立凯. 广藿香FPPS重组蛋白表达及互作蛋白筛选分析[J]. 生物技术通报, 2019, 35(12): 10-15. |
| [13] | 贾建磊, 陈倩, 靳继鹏, 袁赞, 张利平. 绵羊BMPR1B基因真核表达及产物互作蛋白的鉴定[J]. 生物技术通报, 2019, 35(12): 94-104. |
| [14] | 王艺桥,赵新杰,牛芳芳,郭小华,杨博,江元清. 拟南芥中WRKY31转录因子的转录活性与互作蛋白分析[J]. 生物技术通报, 2018, 34(5): 101-109. |
| [15] | 李书鹏, 杨秀芬, 袁京京, 邱德文. 蛋白激发子Hrip1互作蛋白的筛选及其原核表达[J]. 生物技术通报, 2017, 33(6): 182-189. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||