








生物技术通报 ›› 2025, Vol. 41 ›› Issue (4): 176-187.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0903
• 研究报告 • 上一篇
收稿日期:2024-09-19
出版日期:2025-04-26
发布日期:2025-04-25
通讯作者:
梁美霞,女,博士,教授,研究方向 :果树种质资源评价及抗逆育种;E-mail: mxliangdd@163.com作者简介:王天禧,女,硕士研究生,研究方向 :果树学;E-mail: 2864509389@qq.com
基金资助:
WANG Tian-xi( ), YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia(
), YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia( )
)
Received:2024-09-19
Published:2025-04-26
Online:2025-04-25
摘要:
目的 从苹果全基因组中鉴定MdPLATZs转录因子家族,分析其表达特点,为后续深入研究MdPLATZs转录因子的生物学功能奠定基础。 方法 通过生物信息学方法从苹果全基因组鉴定出17个PLATZ基因家族成员,并依据其序列特征将其划分为4个亚家族,并对其理化特征、系统进化关系、基因结构、染色体定位和启动子顺式作用元件等进行分析。本研究测定了苹果不同组织中PLATZ家族成员的表达水平,并通过异源表达及干旱胁迫处理来鉴定MdPLATZ9的基因功能。 结果 生物信息学分析结果表明,MdPLATZs分子量范围为17.23-29.39 kD,等电点介于8.24-9.51之间,且多数成员定位在细胞核内。这些基因分布于苹果的13条染色体上,同一亚家族的成员具有相似的基因结构并具有较高的序列同源性,且各组之间存在明显的共线性关系。PLATZ结构域在MdPLATZs家族中高度保守。基因表达模式分析显示,MdPLATZs家族成员存在组织表达特异性,MdPLATZs在花中均有表达,MdPLATZ7在花和茎中表达量最高。进一步分析显示,过表达MdPLATZ9的拟南芥植株过氧化物酶和过氧化氢酶活性显著增强,同时丙二醛和过氧化氢含量降低,抗坏血酸水平升高,并且超氧阴离子的生成速率减缓,这些结果表明,过表达MdPLATZ9提高了拟南芥的抗旱性。 结论 从苹果全基因组中鉴定出17个MdPLATZs转录因子家族成员,该家族成员在基因结构、蛋白基序表现出高度的相似性和同源性,并且存在显著的片段重复。过表达MdPLATZ9的拟南芥植株表现出明显的抗旱性。
王天禧, 杨炳松, 潘荣君, 盖文贤, 梁美霞. 苹果PLATZ基因家族鉴定及MdPLATZ9基因功能研究[J]. 生物技术通报, 2025, 41(4): 176-187.
WANG Tian-xi, YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia. Identification of the Apple PLATZ Gene Family and Functional Study of the MdPLATZ9 Gene[J]. Biotechnology Bulletin, 2025, 41(4): 176-187.
| 基因ID Gene ID | 基因名称 Gene name | 等电点 pI | 分子量 Molecular weight/Da | 亚细胞定位预测 Localization prediction |
|---|---|---|---|---|
| MD00G1059100 | MdPLATZ1 | 8.55 | 28 544.59 | Nucleus |
| MD02G1017000 | MdPLATZ2 | 8.56 | 29 394.72 | Nucleus |
| MD02G1208800 | MdPLATZ3 | 8.82 | 24 671.44 | Nucleus |
| MD03G1129200 | MdPLATZ4 | 9.51 | 21 971.14 | Nucleus |
| MD05G1248500 | MdPLATZ5 | 8.39 | 28 487.59 | Chloroplast |
| MD06G1001300 | MdPLATZ6 | 8.78 | 28 441.46 | Chloroplast, nucleus |
| MD06G1035500 | MdPLATZ7 | 9.25 | 24 878.17 | Nucleus |
| MD07G1117000 | MdPLATZ8 | 8.81 | 27 385.73 | Nucleus |
| MD10G1229000 | MdPLATZ9 | 8.39 | 28 548.56 | Chloroplast, nucleus |
| MD11G1151300 | MdPLATZ10 | 9.30 | 24 312.98 | Nucleus |
| MD12G1103000 | MdPLATZ11 | 8.88 | 22 043.09 | Nucleus |
| MD13G1017800 | MdPLATZ12 | 9.50 | 24 708.19 | Nucleus |
| MD14G1097400 | MdPLATZ13 | 8.52 | 21 996.09 | Nucleus |
| MD15G1161500 | MdPLATZ14 | 8.75 | 29 101.53 | Nucleus |
| MD16G1015800 | MdPLATZ15 | 9.02 | 25 758.81 | Nucleus |
| MD16G1273400 | MdPLATZ16 | 9.33 | 25 264.61 | Nucleus |
| MD17G1254800 | MdPLATZ17 | 8.24 | 17 230.53 | Nucleus |
表1 苹果MdPLATZ基因家族成员信息
Table 1 Information of MdPLATZ gene family member in apple
| 基因ID Gene ID | 基因名称 Gene name | 等电点 pI | 分子量 Molecular weight/Da | 亚细胞定位预测 Localization prediction |
|---|---|---|---|---|
| MD00G1059100 | MdPLATZ1 | 8.55 | 28 544.59 | Nucleus |
| MD02G1017000 | MdPLATZ2 | 8.56 | 29 394.72 | Nucleus |
| MD02G1208800 | MdPLATZ3 | 8.82 | 24 671.44 | Nucleus |
| MD03G1129200 | MdPLATZ4 | 9.51 | 21 971.14 | Nucleus |
| MD05G1248500 | MdPLATZ5 | 8.39 | 28 487.59 | Chloroplast |
| MD06G1001300 | MdPLATZ6 | 8.78 | 28 441.46 | Chloroplast, nucleus |
| MD06G1035500 | MdPLATZ7 | 9.25 | 24 878.17 | Nucleus |
| MD07G1117000 | MdPLATZ8 | 8.81 | 27 385.73 | Nucleus |
| MD10G1229000 | MdPLATZ9 | 8.39 | 28 548.56 | Chloroplast, nucleus |
| MD11G1151300 | MdPLATZ10 | 9.30 | 24 312.98 | Nucleus |
| MD12G1103000 | MdPLATZ11 | 8.88 | 22 043.09 | Nucleus |
| MD13G1017800 | MdPLATZ12 | 9.50 | 24 708.19 | Nucleus |
| MD14G1097400 | MdPLATZ13 | 8.52 | 21 996.09 | Nucleus |
| MD15G1161500 | MdPLATZ14 | 8.75 | 29 101.53 | Nucleus |
| MD16G1015800 | MdPLATZ15 | 9.02 | 25 758.81 | Nucleus |
| MD16G1273400 | MdPLATZ16 | 9.33 | 25 264.61 | Nucleus |
| MD17G1254800 | MdPLATZ17 | 8.24 | 17 230.53 | Nucleus |
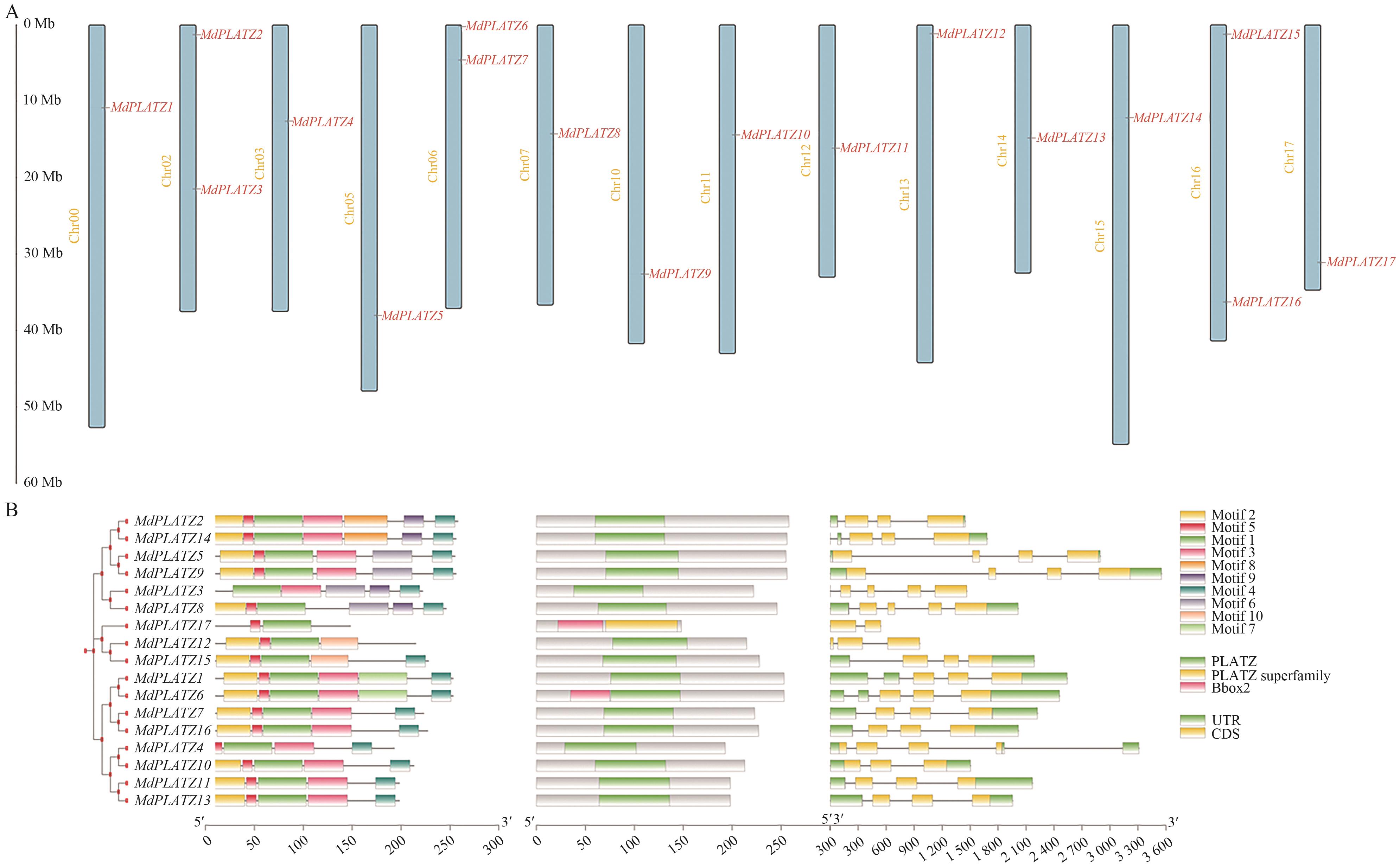
图1 MdPLATZs家族成员的基因染色体定位(A)、基因结构和保守基序分析(B)
Fig. 1 Chromosomal location of genes (A), gene structure and conserved motif analysis (B) of MdPLATZs family members

图2 苹果中PLATZ基因的同源性分析灰色曲线表示苹果基因组中的共线区域,红色曲线表示发生片段重复的基因对
Fig. 2 Homology analysis of PLATZ genes in appleGrey curves indicate regions of colinearity in the apple genome, and red curves indicate pairs of genes where segmental duplication occurs

图3 苹果、拟南芥和玉米的PLATZ系统发育树▲表示苹果PLATZ蛋白;★表示玉米PLATZ蛋白;●表示拟南芥PLATZ蛋白;数字代表亲缘关系的远近
Fig. 3 PLATZ phylogenetic tree of apple, Arabidopsis and maize▲ indicates apple's PLATZ protein. ★ indicates maize's PLATZ protein. ● indicates Arabidopsis's PLATZprotein. Numbers indicate the closeness of kinship

图4 苹果MdPLATZs基因在不同组织表达水平WS:整株幼苗;FR:果实;L:叶;SA:茎尖;FL:花;S:茎;EDB:早期休眠芽;FDB:盛开芽;YL:幼叶
Fig. 4 Expressions of MdPLATZs gene in different tissues of appleWS: Whole seedling; FR: fruit; L: leaf; SA: stem tip; FL: flower; S: stem; EDB: early dormant bud; FDB: bloom bud; YL: young

图5 MdPLATZs基因启动子中顺式作用元件分析MdPLATZs启动子种类及数量可视化
Fig. 5 Analysis of cis-acting elements in the promoter of the MdPLATZs geneVisualized types and number of MdPLATZs promoters

图6 MdPLATZs与ORESARA15进化关系图中数字分别代表亲缘关系的远近以及分支长度
Fig. 6 Evolutionary relationship between MdPLATZs and ORESARA15The numbers in the figure indicate the proximity of the relatives and the branch lengths, respectively
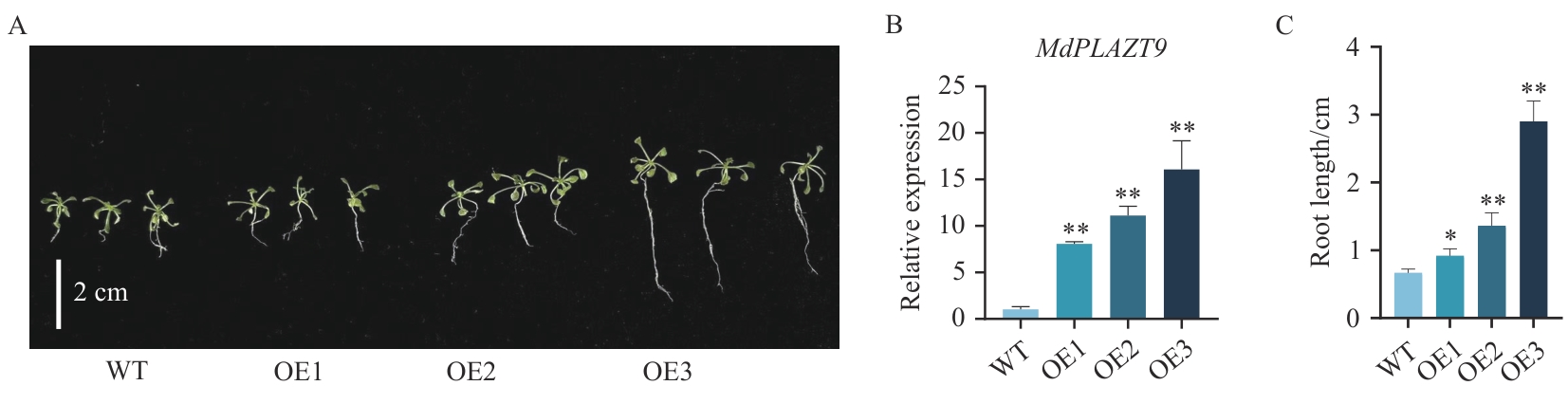
图7 转基因拟南芥与野生型拟南芥的比较A:野生型拟南芥与过表达拟南芥生长14 d表型;B:野生型拟南芥与过表达拟南芥MdPLATZ9表达量;C:野生型拟南芥与过表达拟南芥生长14 d后根长数据统计;WT:野生型拟南芥;OE1:过表达拟南芥株系;OE2:过表达株系2;OE3:过表达株系3;下同;数据为平均值±标准差,n=3;* P<0.05,** P<0.01,*** P<0.001
Fig. 7 Comparison of transgenic A. thaliana with wild-type A. thalianaA: Phenotypes of wild-type Arabidopsisthaliana and overexpressed A. thaliana grown for 14 d. B: Expression of MdPLATZ9 in wild-type A. thaliana and overexpressed A. thaliana. C: Statistics of root length data of wild-type A. thaliana and overexpressed A. thaliana grown for 14 d. WT: Wild-type A. thaliana; OE1: overexpressed A. thaliana line; OE2: overexpressed line 2; OE3: overexpressed line 3; the same below. The data are means±standard deviation (SD), n=3. * P<0.05; ** P<0.01, *** P<0.001

图8 干旱胁迫下野生型与过表达株系拟南芥的表型及生理指标测定A:野生型拟南芥与过表达拟南芥在干旱胁迫下的表型; B‒G:野生型拟南芥与过表达拟南芥在干旱胁迫下O2-生成速率、H2O2含量、CAT活性、POD活性、Vc活性、MDA含量。数据为3个重复样本的平均值±标准偏差,不同字母表示在P<0.05水平差异显著。下同
Fig. 8 Determination of phenotypic and physiological parameters of wild-type and overexpressed strains of A. thaliana under drought stressA: Phenotypes of wild-type A. thaliana and overexpressed A. thaliana under drought stress. B‒G: Rate of O2- production, H2O2 content, CAT activity, POD activity, Vc activity, and MDA content of wild-type A. thaliana and overexpressed A. thaliana under drought stress. Data are the mean±SD of three replicates. Different letters indicate significant differences at P<0.05 level. The same below
| 1 | Strader L, Weijers D, Wagner D. Plant transcription factors-being in the right place with the right company [J]. Curr Opin Plant Biol, 2022, 65: 102136. |
| 2 | Zhang KM, Lan YG, Shi YN, et al. Systematic analysis and functional characterization of the PLATZ transcription factors in moso bamboo (Phyllostachys edulis) [J]. J Plant Growth Regul, 2023, 42(1): 218-236. |
| 3 | Nagano Y, Furuhashi H, Inaba T, et al. A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences [J]. Nucleic Acids Res, 2001, 29(20): 4097-4105. |
| 4 | González-Morales SI, Chávez-Montes RA, Hayano-Kanashiro C, et al. Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana [J]. Proc Natl Acad Sci U S A, 2016, 113(35): E5232-E5241. |
| 5 | Kim JH, Kim J, Jun SE, et al. ORESARA15, a PLATZ transcription factor, mediates leaf growth and senescence in Arabidopsis [J]. New Phytol, 2018, 220(2): 609-623. |
| 6 | Dong T, Yin XM, Wang HT, et al. ABA-INDUCED expression 1 is involved in ABA-inhibited primary root elongation via modulating ROS homeostasis in Arabidopsis [J]. Plant Sci, 2021, 304: 110821. |
| 7 | Li Q, Wang JC, Ye JW, et al. The maize imprinted gene floury3 encodes a PLATZ protein required for tRNA and 5S rRNA transcription through interaction with RNA polymerase III [J]. Plant Cell, 2017, 29(10): 2661-2675. |
| 8 | Li H, Wang YY, Xiao QL, et al. Transcription factor ZmPLATZ2 positively regulate the starch synthesis in maize [J]. Plant Growth Regul, 2021, 93(3): 291-302. |
| 9 | Wang AH, Hou QQ, Si LZ, et al. The PLATZ transcription factor GL6 affects grain length and number in rice [J]. Plant Physiol, 2019, 180(4): 2077-2090. |
| 10 | Zhou SR, Xue HW. The rice PLATZ protein SHORT GRAIN6 determines grain size by regulating spikelet hull cell division [J]. J Integr Plant Biol, 2020, 62(6): 847-864. |
| 11 | Azim JB, Khan MFH, Hassan L, et al. Genome-wide characterization and expression profiling of plant-specific PLATZ transcription factor family genes in Brassica rapa L. [J]. Plant Breed Biotech, 2020, 8(1): 28-45. |
| 12 | Wang JC, Ji C, Li Q, et al. Genome-wide analysis of the plant-specific PLATZ proteins in maize and identification of their general role in interaction with RNA polymerase III complex [J]. BMC Plant Biol, 2018, 18(1): 221. |
| 13 | Fu YX, Cheng MP, Li ML, et al. Identification and characterization of PLATZ transcription factors in wheat [J]. Int J Mol Sci, 2020, 21(23): 8934. |
| 14 | Zhang SC, Yang R, Huo YQ, et al. Expression of cotton PLATZ1 in transgenic Arabidopsis reduces sensitivity to osmotic and salt stress for germination and seedling establishment associated with modification of the abscisic acid, gibberellin, and ethylene signalling pathways [J]. BMC Plant Biol, 2018, 18(1): 218. |
| 15 | Blair EJ, Bonnot T, Hummel M, et al. Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis [J]. Sci Rep, 2019, 9(1): 4814. |
| 16 | So H, Choi SJ, Chung E, et al. Molecular characterization of stress-inducible PLATZ gene from soybean (Glycine max L.) [J]. Plant Omics, 2015, 8(6):479-484. |
| 17 | Liu SS, Yang R, Liu M, et al. PLATZ2 negatively regulates salt tolerance in Arabidopsis seedlings by directly suppressing the expression of the CBL4/SOS3 and CBL10/SCaBP8 genes [J]. J Exp Bot, 2020, 71(18): 5589-5602. |
| 18 | Zenda T, Liu ST, Wang X, et al. Key maize drought-responsive genes and pathways revealed by comparative transcriptome and physiological analyses of contrasting inbred lines [J]. Int J Mol Sci, 2019, 20(6): 1268. |
| 19 | Zhao JY, Zheng L, Wei JT, et al. The soybean PLATZ transcription factor GmPLATZ17 suppresses drought tolerance by interfering with stress-associated gene regulation of GmDREB5 [J]. Crop J, 2022, 10(4): 1014-1025. |
| 20 | Qi JH, Wang H, Wu XY, et al. Genome-wide characterization of the PLATZ gene family in watermelon (Citrullus lanatus L.) with putative functions in biotic and abiotic stress response [J]. Plant Physiol Biochem, 2023, 201: 107854. |
| 21 | Starkus A, Morkūnaitė-Haimi Š, Gurskas T, et al. The biological and genetic mechanisms of fruit drop in apple tree (Malus × domestica Borkh.) [J]. Horticulturae, 2024, 10(9): 987. |
| 22 | Urrestarazu J, Muranty H, Denancé C, et al. Genome-wide association mapping of flowering and ripening periods in apple [J]. Front Plant Sci, 2017, 8: 1923. |
| 23 | Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0 [J]. Mol Biol Evol, 2013, 30(12): 2725-2729. |
| 24 | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| 25 | Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana [J]. Plant J, 1998, 16(6): 735-743. |
| 26 | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method [J]. Nat Protoc, 2008, 3: 1101-1108. |
| 27 | Meng D, Li YY, Bai Y, et al. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress [J]. Plant Physiol Biochem, 2016, 103: 71-83. |
| 28 | Huang C, Yu QB, Lv RH, et al. The reduced plastid-encoded polymerase-dependent plastid gene expression leads to the delayed greening of the Arabidopsis fln2 mutant [J]. PLoS One, 2013, 8(9): e73092. |
| 29 | Farinati S, Rasori A, Varotto S, et al. Rosaceae fruit development, ripening and post-harvest: an epigenetic perspective [J]. Front Plant Sci, 2017, 8: 1247. |
| 30 | Velasco R, Zharkikh A, Affourtit J, et al. The genome of the domesticated apple (Malus × domestica Borkh.) [J]. Nat Genet, 2010, 42(10): 833-839. |
| 31 | Han XL, Liu K, Yuan GP, et al. Genome-wide identification and characterization of AINTEGUMENTA-LIKE (AIL) family genes in apple (Malus domestica Borkh.) [J]. Genomics, 2022, 114(2): 110313. |
| 32 | Meyers BC, Kozik A, Griego A, et al. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis [J]. Plant Cell, 2003, 15(4): 809-834. |
| 33 | Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome [J]. Science, 2001, 291(5507): 1304-1351. |
| 34 | Timilsina R, Kim Y, Park S, et al. ORESARA15, a PLATZ transcription factor, controls root meristem size through auxin and cytokinin signalling-related pathways [J]. J Exp Bot, 2022, 73(8): 2511-2524. |
| 35 | Cai KF, Song XJ, Yue WH, et al. Identification and functional characterization of abiotic stress tolerance-related PLATZ transcription factor family in barley (Hordeum vulgare L.) [J]. Int J Mol Sci, 2024, 25(18): 10191. |
| 36 | Zhang KM, Lan YG, Wu M, et al. PhePLATZ1, a PLATZ transcription factor in moso bamboo (Phyllostachys edulis), improves drought resistance of transgenic Arabidopsis thaliana [J]. Plant Physiol Biochem, 2022, 186: 121-134. |
| 37 | Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana [J]. Plant J, 2005, 43(1): 68-78. |
| 38 | 冯军, 郑彩霞. DREB转录因子在植物非生物胁迫中的作用及应用研究 [J]. 植物生理学报, 2011, 47(5): 437-442. |
| Feng J, Zheng CX. Research and application prospect of DREB transcription factor in plant abiotic stress resistance [J]. Plant Physiol J, 2011, 47(5): 437-442. | |
| 39 | Chen YF, Li Z, Sun TT, et al. Sugarcane ScDREB2B-1 confers drought stress tolerance in transgenic Nicotiana benthamiana by regulating the ABA signal, ROS level and stress-related gene expression[J]. Int J Mol Sci,2022, 23(17): 9557. |
| [1] | 刘涛, 王志淇, 吴文博, 石文婷, 王超楠, 杜崇, 杨中敏. 马铃薯GRAM基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(4): 145-155. |
| [2] | 杨朝结, 张兰, 陈红, 黄娟, 石桃雄, 朱丽伟, 陈庆富, 李洪有, 邓娇. 苦荞转录因子基因FtbHLH3调控类黄酮生物合成的功能鉴定[J]. 生物技术通报, 2025, 41(4): 134-144. |
| [3] | 王田田, 常雪瑞, 黄婉洋, 黄嘉欣, 苗如意, 梁燕平, 王静. 辣椒GASA基因家族的鉴定及分析[J]. 生物技术通报, 2025, 41(4): 166-175. |
| [4] | 张益瑄, 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰. 马铃薯DIR家族全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(3): 123-136. |
| [5] | 覃悦, 杨妍, 张磊, 卢丽丽, 李先平, 蒋伟. 二倍体和四倍体马铃薯StGAox基因鉴定与比较分析[J]. 生物技术通报, 2025, 41(3): 146-160. |
| [6] | 王琛, 刘国梅, 陈畅, 张晋龙, 姚琳, 孙璇, 杜春芳. 白菜型油菜CCDs家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(3): 161-170. |
| [7] | 韩江涛, 张帅博, 秦雅蕊, 韩硕洋, 张雅康, 王吉庆, 杜清洁, 肖怀娟, 李猛. 甜瓜β-淀粉酶基因家族的鉴定及对非生物胁迫的响应[J]. 生物技术通报, 2025, 41(3): 171-180. |
| [8] | 宋姝熠, 蒋开秀, 刘欢艳, 黄亚成, 刘林娅. ‘红阳’猕猴桃TCP基因家族鉴定及其在果实中的表达分析[J]. 生物技术通报, 2025, 41(3): 190-201. |
| [9] | 王斌, 王玉昆, 肖艳辉. 丁香罗勒(Ocimum gratissimum)叶片响应镉胁迫的比较转录组学分析[J]. 生物技术通报, 2025, 41(3): 255-270. |
| [10] | 刘洁, 王飞, 陶婷, 张玉静, 陈浩婷, 张瑞星, 石玉, 张毅. 过表达SlWRKY41提高番茄幼苗抗旱性[J]. 生物技术通报, 2025, 41(2): 107-118. |
| [11] | 颜伟, 陈慧婷, 叶青, 刘广超, 刘新, 侯丽霞. 葡萄HCT基因家族鉴定及其对低温胁迫的响应[J]. 生物技术通报, 2025, 41(2): 175-186. |
| [12] | 李艳伟, 杨妍妍, 孙亚玲, 霍雨猛, 王振宝, 刘冰江. 基于转录组分析植物激素对洋葱鳞茎膨大发育的调控机制[J]. 生物技术通报, 2025, 41(2): 187-201. |
| [13] | 匡健华, 程志鹏, 赵永晶, 杨洁, 陈润乔, 陈龙清, 胡慧贞. 激素和非生物胁迫下荷花GH3基因家族的表达分析[J]. 生物技术通报, 2025, 41(2): 221-233. |
| [14] | 钱政毅, 吴绍芳, 曹舒怡, 宋雅欣, 潘鑫峰, 李兆伟, 范凯. 睡莲NAC转录因子的鉴定及其表达分析[J]. 生物技术通报, 2025, 41(2): 234-247. |
| [15] | 黄颖, 遇文婧, 刘雪峰, 刁桂萍. 山新杨谷胱甘肽转移酶基因的生物信息学与表达模式分析[J]. 生物技术通报, 2025, 41(2): 248-256. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||