








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (8): 275-283.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1247
Previous Articles Next Articles
HE Xiao-li1,3( ), GUO Lei-zhou1,2, HAN Jia-hui2, TANG Yin1,2, YUAN Yuan1,2, DAI Qi-lin1, PING Shu-zhen2, JIANG Shi-jie1(
), GUO Lei-zhou1,2, HAN Jia-hui2, TANG Yin1,2, YUAN Yuan1,2, DAI Qi-lin1, PING Shu-zhen2, JIANG Shi-jie1( )
)
Received:2020-10-11
Online:2021-08-26
Published:2021-09-10
Contact:
JIANG Shi-jie
E-mail:927267653@qq.com;sjjiang0406@swust.edu.cn
HE Xiao-li, GUO Lei-zhou, HAN Jia-hui, TANG Yin, YUAN Yuan, DAI Qi-lin, PING Shu-zhen, JIANG Shi-jie. Research Progress on Bacterial Periplasmic Chaperone LolA[J]. Biotechnology Bulletin, 2021, 37(8): 275-283.

Fig. 1 Crystal structure of Escherichia coli LolA[22] The hydrophobic cavity of LolA is composed of an open β-barrel and an α-helical lid,which is also a binding site for lipids;the C-terminal loop of LolA is composed of a short α-helix and a twelfth β-strand,which can correctly combine lipoprotein delivery to OM

Fig. 2 Lipoprotein transport and outer membrane anchoring mediated by the Lol system[22] a:Lipoprotein is first placed in LolE(1),and then transferred to LolA located on LolC(2). When LolCDE binds to lipoproteins,the interaction between LolA and LolCDE increases(solid arrow). Then LolA forms a hydrophilic complex with lipoproteins in an ATP-dependent manner,and opens the hydrophobic cavity of LolA at the same time(3). b:After passing through the periplasmic space,LolA and LolB interact in a “mouth to mouth” manner to transfer lipoproteins from LolA to LolB(4),and finally locate to the outer membrane
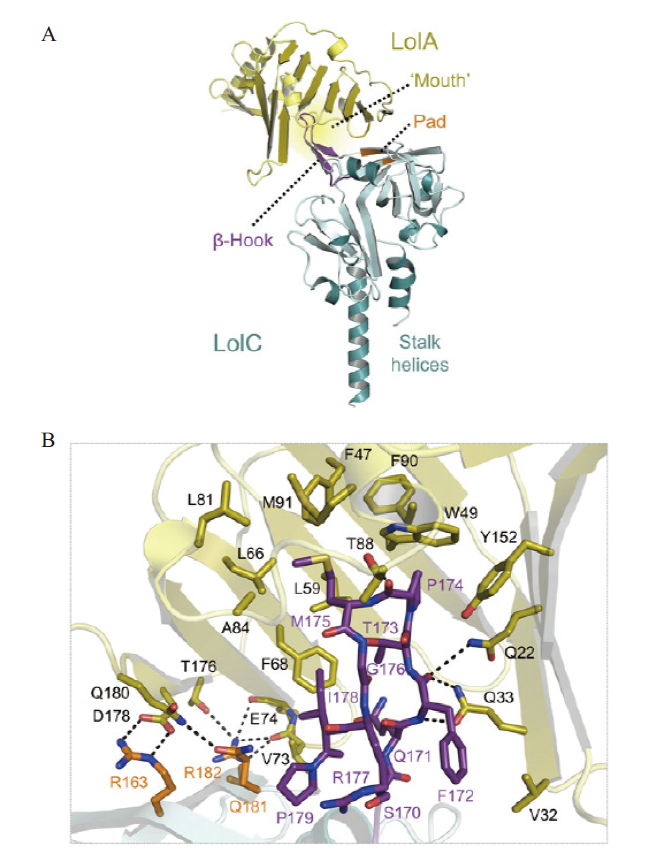
Fig. 3 Crystal structure of LolA is combined with the periplasmic domain of LolC[30] A:The overall structure of the LolA-LolC complex. B:Interaction interface view. LolC and LolA are represented in cyan and gold,respectively. The residue belonging to Hook and Pad is shown in purple and orange. The content that interacts with LolC is displayed in a bar shape
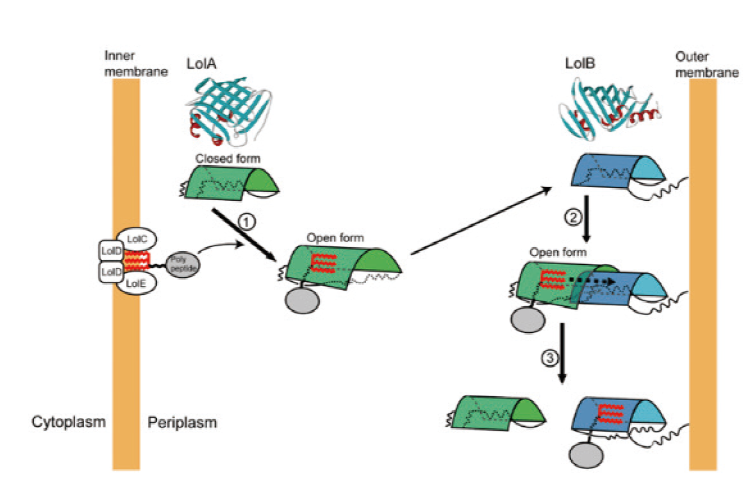
Fig. 4 Transport model of lipoprotein on LolA and LolB[36] LolA and LolB are represented by green and blue barrels,respectively,and red represents the acyl chain. Step 1: a lipoprotein is obtained from LolCDE on the periplasmic side of the inner membrane. LolA undergoes a conformational change and adapts its hydrophobic cavity to 1 to 3 lipoprotein acyl chains. Step 2: the lipoprotein-LolA complex interacts with the LolB anchored on the outer membrane. In the complex,LolA and LolB form a hydrophobic,channel-like structure. Step 3: the acyl chain of the lipoprotein is transferred from LolA to LolB through the hydrophobic channel formed by these two proteins

Fig.5 Binding mode of BLP in the hydrophobic cavity of LolA[47] A:Left:Start configuration,where BLP(yellow)is located outside the LolA(cyan)cavity. Upper right:The combination of the three fat tails of BLP near the mouth of the cavity. Lower right:One fat tail is deep in the cavity,and the other two are at the mouth of the cavity. B:The minimum distance between the three independently simulated BLP fat tails and F90 residues(as shown in the illustration by Van der Waals notation)
| [1] |
Sankaran K, Wu HC. Lipid modification of bacterial Prolipoprotei. Transfer of diacylglyceryl moiety from phosphatidylglycerol[J]. Journal of Biological Chemistry, 1994, 269(31):19701-19706.
pmid: 8051048 |
| [2] |
Buddelmeijer N. The molecular mechanism of bacterial lipoprotein modification--how, when and why?[J]. FEMS Microbiol Rev 2015, 39(2):246-261.
doi: 10.1093/femsre/fuu006 pmid: 25670733 |
| [3] |
Lovullo ED, Wright LF, Isabella V, et al. Revisiting the Gram-negative lipoprotein paradigm[J]. J Bacteriol, 2015, 197(10):1705-1715.
doi: 10.1128/JB.02414-14 URL |
| [4] |
Hutchings MI, Palmer T, Harrington DJ, et al. Lipoprotein biogenesis in Gram-positive bacteria:knowing when to hold'em, knowing when to fold 'em[J]. Trends Microbiol, 2009, 17(1):13-21.
doi: 10.1016/j.tim.2008.10.001 pmid: 19059780 |
| [5] |
Nakayama H, Kurokawa K, Lee BL. Lipoproteins in bacteria:structures and biosynthetic pathways[J]. FEBS J, 2012, 279(23):4247-4268.
doi: 10.1111/febs.12041 pmid: 23094979 |
| [6] |
Schenk M, Belisle JT, Modlin RL. TLR2 looks at lipoproteins[J]. Immunity, 2009, 31(6):847-849.
doi: 10.1016/j.immuni.2009.11.008 URL |
| [7] |
Bernadac A, Gavioli M, Lazzaroni JC, et al. Escherichia coli tol-pal mutants form outer membrane vesicles[J]. J Bacteriol, 1998, 180:4872-4878.
pmid: 9733690 |
| [8] |
Clavel T, Germon P, Vianney A, et al. Tol B protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA[J]. Mol Microbiol, 1998, 29:359-367.
pmid: 9701827 |
| [9] |
Ehrmann M, Ehrle R, Hofmann E, et al. The ABC maltose transporter[J]. Mol Microbiol, 1998, 29:685-694.
pmid: 9723909 |
| [10] |
Nikaido H. Multiple antibiotic resistance and efflux[J]. Curr Opin Microbiol, 1998, 1:516-523.
pmid: 10066525 |
| [11] |
Tokuda H. Biogenesis of outer membranes in Gram-negative bacteria[J]. Bioscience, Biotechnology, and Biochemistry, 2009, 73(3):465-473.
doi: 10.1271/bbb.80778 URL |
| [12] |
Doerrler WT. Lipid trafficking to the outer membrane of Gram-negative bacteria[J]. Molecular Microbiology, 2006, 60(3):542-552.
pmid: 16629659 |
| [13] | Lorenz C, Dougherty TJ, Lory S. Correct sorting of lipoproteins into the inner and outer membranes of Pseudomonas aeruginosa by the Escherichia coli LolCDE transport system[J]. mBio, 2019, 10(2):e00194-19. |
| [14] | Narita S, Tokuda H. Bacterial lipoproteins;biogenesis, sorting and quality control[J]. Biochimica et Biophysica Acta(BBA)-Molec-ular and Cell Biology of Lipids, 2017, 1862(11):1414-1423. |
| [15] | Zückert WR. Secretion of bacterial lipoproteins:through the cytoplasmic membrane, the periplasm and beyond[J]. Biochimica et Biophysica Acta(BBA)-Molecular Cell Research, 2014, 1843(8):1509-1516. |
| [16] |
Matsuyama S, Tajima T, Tokuda H. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane[J]. The EMBO Journal, 1995, 14(14):3365.
doi: 10.1002/embj.1995.14.issue-14 URL |
| [17] |
Konovalova A, Silhavy TJ. Outer membrane lipoprotein biogenesis:Lol is not the end[J]. Philos Trans R Soc Lond B Biol Sci, 2015, 370:20150030.
doi: 10.1098/rstb.2015.0030 URL |
| [18] | Goemans C, Denoncin K, Collet JF. Folding mechanisms of periplasmic proteins[J]. Biochimica et Biophysica Acta(BBA)-Molecular Cell Research, 2014, 1843(8):1517-1528. |
| [19] |
Takeda K, Miyatake H, Yokota N, et al. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB[J]. EMBO J, 2003, 22:3199-3209.
doi: 10.1093/emboj/cdg324 URL |
| [20] |
Okuda S, Watanabe S, Tokuda H. A short helix in the C-terminal region of LolA is important for the specific membrane localization of lipoproteins[J]. FEBS Lett, 2008, 582:2247-2251.
doi: 10.1016/j.febslet.2008.05.022 URL |
| [21] |
Murahari P, Anishetty S, Pennathur G, et al. Research article:Understanding the lid movements of LolA in Escherichia coli using molecular dynamics simulation and in silico point mutation[J]. Computational Biology and Chemistry, 2013, 47:71-80.
doi: 10.1016/j.compbiolchem.2013.06.005 URL |
| [22] |
Okuda S, Tokuda H. Lipoprotein Sorting in Bacteria[J]. Annu Rev Microbiol, 2011, 65(1):239-259.
doi: 10.1146/annurev-micro-090110-102859 URL |
| [23] |
Pastukhov AV, Ropson IJ. Fluorescent dyes as probes to study lipid-binding proteins[J]. Proteins, 2003, 53:607-615.
doi: 10.1002/(ISSN)1097-0134 URL |
| [24] |
Oguchi Y, Takeda K, Watanabe S, et al. Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release[J]. J Biol Chem, 2008, 283:25414-25420.
doi: 10.1074/jbc.M804736200 pmid: 18617521 |
| [25] |
Tao K, Watanabe S, Narita SI, et al. A periplasmic LolA derivative with a lethal disulfide bond activates the Cpx stress response system[J]. Journal of Bacteriology, 2010, 192(21):5657-5662.
doi: 10.1128/JB.00821-10 URL |
| [26] |
Okuda S, Tokuda H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB[J]. Proceedings of the National Academy of Sciences, 2009, 106(14):5877-5882.
doi: 10.1073/pnas.0900896106 URL |
| [27] |
Mizutani M, Mukaiyama K, Xiao J, et al. Functional differentiation of structurally similar membrane subunits of the ABC transporter LolCDE complex[J]. FEBS letters, 2013, 587(1):23-29.
doi: 10.1016/j.febslet.2012.11.009 URL |
| [28] |
Narita S, Tanaka K, Matsuyama S, et al. Disruption of lolCDE, encoding an ATP-binding cassette transporter, is lethal for Escherichia coli and prevents release of lipoproteins from the inner membrane[J]. Journal of Bacteriology, 2002, 184(5):1417-1422.
doi: 10.1128/JB.184.5.1417-1422.2002 URL |
| [29] |
Yasuda M, Iguchi-Yokoyama A, Matsuyama SI, et al. Membrane topology and functional importance of the periplasmic region of ABC transporter LolCDE[J]. Bioscience, Biotechnology, and Biochemistry, 2009, 73(10):2310-2316.
doi: 10.1271/bbb.90451 URL |
| [30] | Kaplan E, Greene NP, Crow A, et al. Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(31):E7389-E7397. |
| [31] |
Watanabe S, Oguchi Y, Yokota N, et al. Large-scale preparation of the homogeneous LolA-lipoprotein complex and efficient in vitro transfer of lipoproteins to the outer membrane in a LolB-dependent manner[J]. Protein Science, 2007, 16(12):2741-2749.
doi: 10.1110/(ISSN)1469-896X URL pmid: 18029423 |
| [32] |
Ito Y, Kanamaru K, Taniguchi N, et al. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins[J]. Molecular Microbiology, 2006, 62(4):1064-1075.
doi: 10.1111/mmi.2006.62.issue-4 URL |
| [33] |
Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli[J]. Nature methods, 2006, 3(4):263-265.
doi: 10.1038/nmeth864 URL |
| [34] |
Wang L, Xie J, Schultz PG. Expanding the genetic code[J]. Annu Rev Biophys Biomol Struct, 2006, 35:225-249.
doi: 10.1146/annurev.biophys.35.101105.121507 URL |
| [35] |
Szewczyk J, Collet JF. The journey of lipoproteins through the cell:one birthplace, multiple destinations[J]. Adv Microb Physiol, 2016, 69:1-50.
doi: S0065-2911(16)30024-8 pmid: 27720009 |
| [36] |
Nakada S, Sakakura M, Takahashi H, et al. Structural investigation of the interaction between LolA and LolB using NMR[J]. Journal of Biological Chemistry, 2009, 284(36):24634-24643.
doi: 10.1074/jbc.M109.001149 URL |
| [37] |
Matsuyama SI, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB(Hem M), involved in the LolA(p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli[J]. EMBO J, 1997, 16:6947-6955.
pmid: 9384574 |
| [38] |
Yokota N, Kuroda T, Matsuyama S, et al. Characterization of the Lol A-Lol B system as the general lipoprotein localization mechanism of Escherichia coli[J]. J Biol Chem, 1999, 274(43):30995-30999.
pmid: 10521496 |
| [39] |
Masuda K, Matsuyama S, Tokuda H. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization[J]. Proc Natl Acad Sci USA, 2002, 99(11):7390-7395.
doi: 10.1073/pnas.112085599 URL |
| [40] |
Tajima T, Yokota N, Matsuyama S, et al. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins[J]. FEBS Letters, 1998, 439(1-2):51-54.
pmid: 9849875 |
| [41] |
Miyamoto A, Matsuyama S, Tokuda H. Mutant of LolA, a lipoprotein-specific molecular chaperone of Escherichia coli, defective in the transfer of lipoproteins to LolB[J]. Biochem Biophys Res Commun, 2001, 287(5):1125-1128.
doi: 10.1006/bbrc.2001.5705 URL |
| [42] |
Tao K, Narita S, Tokuda H, et al. Defective lipoprotein sorting induces lolA expression through the rcs stress response phosphorelay system[J]. Journal of Bacteriology, 2012, 194(14):3643-3650.
doi: 10.1128/JB.00553-12 URL |
| [43] |
Miyamoto A, Matsuyama SI, Tokuda H. Dominant negative mutant of a lipoprotein-specific molecular chaperone, LolA, tightly associates with LolCDE[J]. FEBS Letters, 2002, 528(1/3):193-196.
doi: 10.1016/S0014-5793(02)03305-7 URL |
| [44] |
Grabowicz M, Silhavy TJ. Redefining the essential trafficking pathway for outer membrane lipoproteins[J]. Proc Natl Acad Sci USA, 2017, 114(18):4769-4774.
doi: 10.1073/pnas.1702248114 URL |
| [45] |
Choi U, Lee C. Antimicrobial agents that inhibit the outer membrane assembly machines of gram-negative bacteria[J]. Journal of Microbiology and Biotechnology, 2019, 29(1):1-10.
doi: 10.4014/jmb.1804.03051 URL |
| [46] |
Muheim C, Gotzke H, Eriksson A, et al. Increasing the permeability of Escherichia coli using MAC13243[J]. Scientific Reports, 2017, 7(1):17629.
doi: 10.1038/s41598-017-17772-6 URL |
| [47] |
Boags AT, Samsudin F, Khalid S, et al. Details of hydrophobic entanglement between small molecules and Braun’s lipoprotein within the cavity of the bacterial chaperone LolA[J]. Scientific Reports, 2019, 9(1):3717.
doi: 10.1038/s41598-019-40170-z URL |
| [48] |
Bosch V, Braun V. Distribution of murein-lipoprotein between the cytoplasmic and outer membrane of Escherichia coli[J]. FEBS Lett, 1973, 34(2):307-310.
pmid: 4583850 |
| [49] |
Shu W, Liu J, Ji H, et al. Core structure of the outer membrane lipoprotein from Escherichia coli at 1. 9 A resolution[J]. J Mol Biol, 2000, 299(4):1101-1112.
pmid: 10843861 |
| [50] |
Watanabe S, Oguchi Y, Takeda K, et al. Introduction of a lethal redox switch that controls the opening and closing of the hydrophobic cavity in LolA[J]. Journal of Biological Chemistry, 2008, 283(37):25421-25427.
doi: 10.1074/jbc.M804737200 pmid: 18621730 |
| [51] |
Pathania R, Zlitni S, Barker C, et al. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting[J]. Nat Chem Biol, 2009, 5(11):849-856.
doi: 10.1038/nchembio.221 pmid: 19783991 |
| [52] |
Narita S, Tanaka K, Matsuyama S, et al. Disruption of lolCDE encoding an ATP-binding-cassette transporter is lethal for Escherichia coli and prevents the release of lipoproteins from the inner membrane[J]. J Bacteriol, 2002, 184(5):1417-1422.
doi: 10.1128/JB.184.5.1417-1422.2002 URL |
| [53] |
Juncker AS, Willenbrock H, Von Heijne G, et al. Prediction of lipoprotein signal peptides in gram-negative bacteria[J]. Protein Sci, 2003, 12(8):1652-1662.
doi: 10.1110/ps.0303703 URL |
| [54] |
Lewenza S, Vidal-Ingigliardi D, Pugsley AP. Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae[J]. J Bacteriol, 2006, 188(10):3516-3524.
pmid: 16672606 |
| [55] | Grabowicz M. Lipoproteins and their trafficking to the outer membrane[J]. EcoSal Plus, 2019, 8:0038. |
| [56] |
Schulze R J, Chen SY, Kumru O S, et al. Translocation of Borrelia burgdorferi surface lipoprotein OspA through the outer membrane requires an unfolded conformation and can initiate at the C-terminus[J]. Mol Microbiol, 2010, 76(5):1266-1278.
doi: 10.1111/j.1365-2958.2010.07172.x pmid: 20398211 |
| [57] |
Rhodes RG, Samarasam MN, Van Groll EJ, et al. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion[J]. J Bacteriol, 2011, 193(19):5322-5327.
doi: 10.1128/JB.05480-11 pmid: 21784937 |
| [58] |
Liao CT, Chiang YC, Hsiao YM. Functional characterization and proteomic analysis of lolA in Xanthomonas campestris pv. Campestris[J]. BMC Microbiology, 2019, 19(1):20.
doi: 10.1186/s12866-019-1387-9 URL |
| [1] | DONG Hai-jiao, YANG Xiao-yu, MO Bei-xin, CHEN Xue-mei, CUI Jie. Research Progress in NAD+ Cap Modification at the 5' End of RNA [J]. Biotechnology Bulletin, 2022, 38(2): 245-251. |
| [2] | ZHAO Jie, LI An, LIANG Gang, JIN Xin-xin, PAN Li-gang. Research Progress in the Biological Functions of Plant circRNAs [J]. Biotechnology Bulletin, 2022, 38(10): 1-9. |
| [3] | WANG Zhi-shan, LI Ni, WANG Wei-ping, LIU Yang. Research Progress in Endophytic Bacteria in Rice Seeds [J]. Biotechnology Bulletin, 2022, 38(1): 236-246. |
| [4] | LUO Wei, MU Qiong, SHU Jian-hong, WU Jia-hai, WANG Xiao-li. Expression,Protein Interactions and Biological Function Analysis of FaFT in Festuca arundinacea [J]. Biotechnology Bulletin, 2021, 37(4): 8-17. |
| [5] | XUE Xiang-lan, DING Yang-yang, LIU Yue, LI Xiao-bo, JIANG Lin, HE Xiao-hong, MA Yue-hui, ZHAO Qian-jun. Research Progress on Biological Function Growth and Development Related to N6-methyladenosine in Mammals [J]. Biotechnology Bulletin, 2021, 37(4): 251-259. |
| [6] | FENG Yi-long, ZHANG Wen-li. Research Progress on DNA Guanine Quadruplex [J]. Biotechnology Bulletin, 2020, 36(7): 23-31. |
| [7] | HUANG Xing, DING Feng, PENG Hong-xiang, PAN Jie-chun, HE Xin-hua, XU Jiong-zhi, LI Lin. Research Progress on Family of Plant WRKY Transcription Factors [J]. Biotechnology Bulletin, 2019, 35(12): 129-143. |
| [8] | WANG Chun-yu, ZHANG Qian. Research Progress on Plant NAC Transcription Factors [J]. Biotechnology Bulletin, 2018, 34(11): 8-14. |
| [9] | XU Hai-dong, LENG Qi-ying, PATRICIA Adu-Asiamah, WANG Zhang, LI Ting, ZHANG Li. Circular RNAs:Research Progress and Its Significance in Birds and Livestock [J]. Biotechnology Bulletin, 2018, 34(11): 56-69. |
| [10] | ZHANG Ying-yue ,MA Yue-hui ,ZHAO Qian-jun. Study Progress on Circular RNA [J]. Biotechnology Bulletin, 2017, 33(7): 29-34. |
| [11] | LUO Yan,LIU Xiao-gang,ZHOU Zhi-qin. Research Progress on Methods for Isolating the Gene of Plant Glycosyltransferase,and Its Biological Functions [J]. Biotechnology Bulletin, 2016, 32(12): 34-39. |
| [12] | ZHENG Chao, LI Deng-gao, BAI Wei. Advances on Cysteine-rich Receptor-like Kinases in Plants [J]. Biotechnology Bulletin, 2016, 32(11): 10-17. |
| [13] | Niu Xulong, Feng Wanjun, Ma Jinhu, Xing Guofang. Research Progress on Biological Functions of Long Non-coding RNA in Plants [J]. Biotechnology Bulletin, 2015, 31(6): 1-7. |
| [14] | Jiang Shanshan, Zhang Dan, Kong Xiangpei, Zhou Yan, Li Dequan. Research Progress of Structural Characteristics and Functions of Calcium-dependent Protein Kinases in Plants [J]. Biotechnology Bulletin, 2013, 0(6): 12-19. |
| [15] | Tan Yaqing, Liu Dehu. The Prospect and Development of the Human Acid Fibroblast Growth Factor [J]. Biotechnology Bulletin, 2013, 0(5): 22-27. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||