








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (10): 160-171.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1227
Previous Articles Next Articles
JU Kang-hui1( ), TIAN Xiao-ya1, WANG Li2, CHEN Jing-yu1(
), TIAN Xiao-ya1, WANG Li2, CHEN Jing-yu1( )
)
Received:2024-01-02
Online:2024-10-26
Published:2024-11-20
Contact:
CHEN Jing-yu
E-mail:jkh72552365@163.com;chenjy@cau.edu.cn
JU Kang-hui, TIAN Xiao-ya, WANG Li, CHEN Jing-yu. Cell Programming Technology: Paving the Way for Efficient Cell Factories[J]. Biotechnology Bulletin, 2024, 40(10): 160-171.
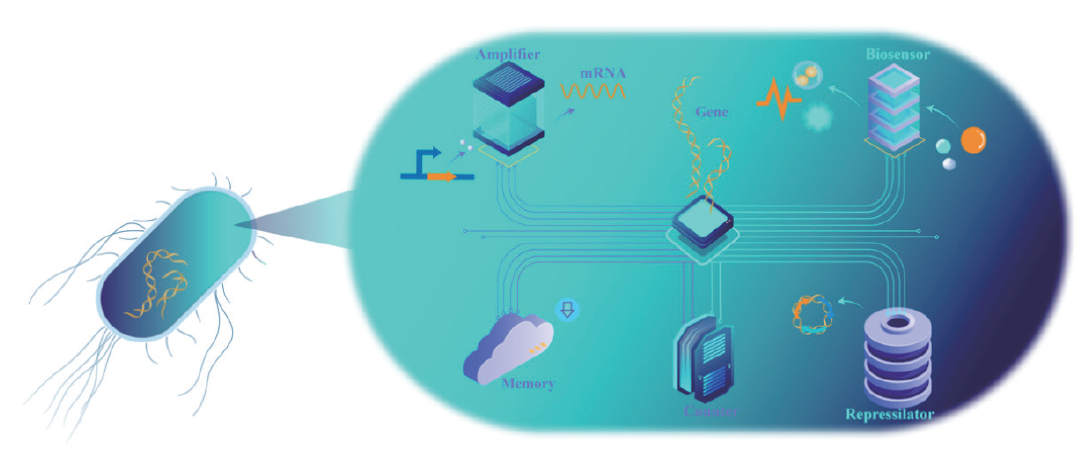
Fig. 1 Topology concepts on cell programming Under genetic regulation, each element assumes a distinct role. Amplifier: It amplifies the transcriptional signals with gain control within cascade gene networks, and functional control inside and outside the cell can be achieved by predicting and dynamically modulating transcriptional signals. Biosensor: Real-time regulation through visual and dynamic monitoring of environmental components both inside and outside the cell factory. Memory: Memory elements within the cell factory extend its working time by preserving essential information. Repressilator: Probing intracellular regulatory dynamics. Counter: Catalyzing complex cell programming, the counter concept facilitates more intricate cell programming and broader biotechnological applications by calculating the cascade reactions of units within the cell
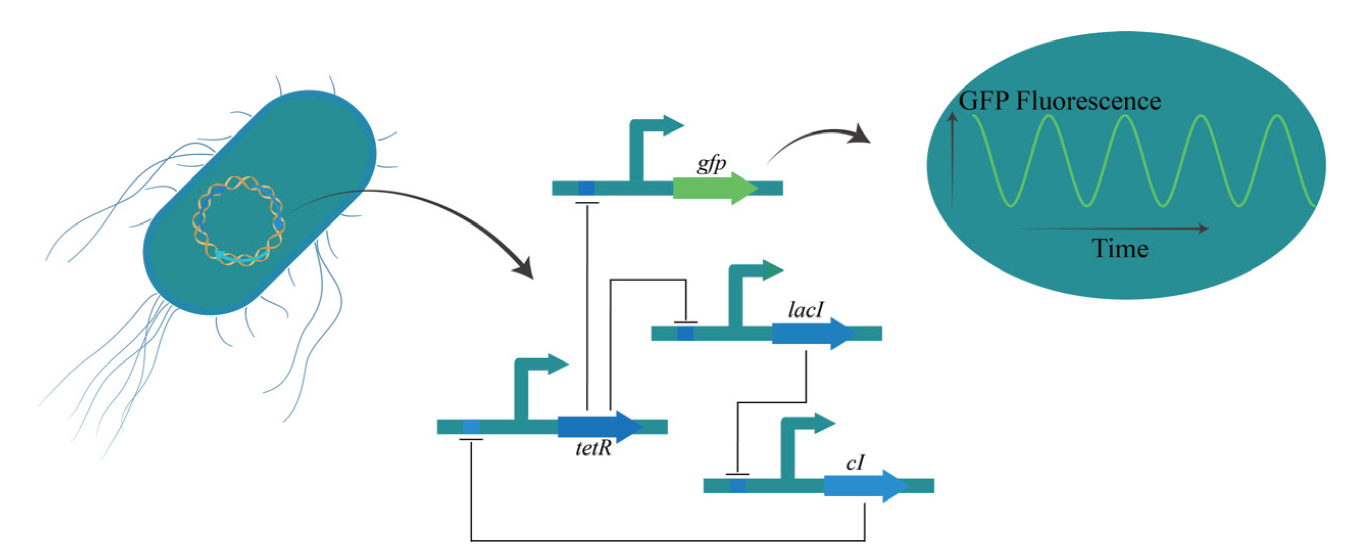
Fig. 2 Work mechanism of initial “repressilator” The “repressilator” comprises three operons interacting and inhibiting with each other. Notably, the tetR gene assumes a pivotal role in governing the regulatory node of GFP. Through the application of a time-lapse fluorescence microscope to monitor the GFP signal, the circuit manifests periodic oscillations
| 细胞编程 Cell reprogramming | 描述 Description | 来源 Reference | |
|---|---|---|---|
| 遗传元件 Genetic elements | 振荡器 | ||
| 抑制振荡器-RFL | 初代“repressilator”,3个转录抑制子建立的振荡网络,开发早、研究范围广、独立控制尚未实现 | [ | |
| 双反馈振荡器-DFO | 由两个诱导操纵子控制,可以通过控制诱导剂的浓度实现振荡器电路的独立控制,使得振荡器能够用于更复杂的电路设计 | [ | |
| 存储器 | |||
| 功能稳态型存储器 | 利用λ噬菌体的Cro蛋白诱导的CI/Cro转录开关作为“记忆元件”使得工程菌能够维持功能稳定至少6个月之久 | [ | |
| 状态机 | 使用重组酶特有的DNA切除和反转功能在大肠杆菌中构建状态机(State machines),读取基因表达的状态,记录所有输入的时序,并对基因表达进行多输入、多输出的控制 | [ | |
| 生物磁带记录器 | 使用基于CRISPR的适应系统,构建了一种“生物磁带记录器”,将其用来描述细胞动态和环境变化,从而相对准确地描述时间间隔生物信号和调节程序的能力 | [ | |
| 生物传感器 | |||
| 产物响应型生物传感器 | 基于产物响应的生物传感器,产物生成量越高,细胞传感荧光水平越强,以此建立了超高通量的筛选方式 | [ | |
| 温敏型生物传感器 | 利用温敏型生物传感器控制启动子表达,解偶联细胞生长与合成的关系,极大地提高了类胡萝卜的产生 | [ | |
| 细胞底盘 Cell chassis | 枯草芽孢杆菌 Bacillus subtilis | 模式化的细胞工厂,生产工业用酶的优良底盘 | [ |
| 酪丁酸梭菌 Clostridium tyrobutyricum | 在生产丁酸及丁醇的生产上有极高的生产效率,短链脂肪酸潜在的理性“细胞工厂” | [ | |
| 米曲霉 Aspergillus oryzae | 在天然产物的生物合成领域有重要的研究价值 | [ | |
| 嗜盐碱单胞菌 Natronomonas kamekura | 用于高分子材料、化学品和燃料的生物制造 | [ | |
| 解脂耶氏酵母 Yarrowia lipolytica | 适合用于油脂及其衍生物的生物合成 | [ | |
| 蓝藻 | 与上转换纳米粒子一起封装,能够创建工程化的微氧细胞工厂 | [ | |
| Minicell | 无染色体细胞,药物递送的优良载体 | [ | |
| Simcell | 无染色体细胞,安全可编程的通用平台 | [ | |
| 复杂基因逻辑回路设计Complex gene logic circuit design | AND、NOT联用型遗传电路设计 | 利用AND和NOT联用的基因电路,巧妙实现了细胞生长过渡到生物合成时间点的精确把控,减少外源电路引入对宿主菌株的代谢负担 | [ |
| CASwitch | 将CRISPR-Cas内环核酸酶与Tet-On3G诱导基因系统相结合而设计的高性能基因电路,用于哺乳动物系统中高性能诱导表达 | [ | |
Table 1 Partial modification basis and technology of the cell programming involved in this study
| 细胞编程 Cell reprogramming | 描述 Description | 来源 Reference | |
|---|---|---|---|
| 遗传元件 Genetic elements | 振荡器 | ||
| 抑制振荡器-RFL | 初代“repressilator”,3个转录抑制子建立的振荡网络,开发早、研究范围广、独立控制尚未实现 | [ | |
| 双反馈振荡器-DFO | 由两个诱导操纵子控制,可以通过控制诱导剂的浓度实现振荡器电路的独立控制,使得振荡器能够用于更复杂的电路设计 | [ | |
| 存储器 | |||
| 功能稳态型存储器 | 利用λ噬菌体的Cro蛋白诱导的CI/Cro转录开关作为“记忆元件”使得工程菌能够维持功能稳定至少6个月之久 | [ | |
| 状态机 | 使用重组酶特有的DNA切除和反转功能在大肠杆菌中构建状态机(State machines),读取基因表达的状态,记录所有输入的时序,并对基因表达进行多输入、多输出的控制 | [ | |
| 生物磁带记录器 | 使用基于CRISPR的适应系统,构建了一种“生物磁带记录器”,将其用来描述细胞动态和环境变化,从而相对准确地描述时间间隔生物信号和调节程序的能力 | [ | |
| 生物传感器 | |||
| 产物响应型生物传感器 | 基于产物响应的生物传感器,产物生成量越高,细胞传感荧光水平越强,以此建立了超高通量的筛选方式 | [ | |
| 温敏型生物传感器 | 利用温敏型生物传感器控制启动子表达,解偶联细胞生长与合成的关系,极大地提高了类胡萝卜的产生 | [ | |
| 细胞底盘 Cell chassis | 枯草芽孢杆菌 Bacillus subtilis | 模式化的细胞工厂,生产工业用酶的优良底盘 | [ |
| 酪丁酸梭菌 Clostridium tyrobutyricum | 在生产丁酸及丁醇的生产上有极高的生产效率,短链脂肪酸潜在的理性“细胞工厂” | [ | |
| 米曲霉 Aspergillus oryzae | 在天然产物的生物合成领域有重要的研究价值 | [ | |
| 嗜盐碱单胞菌 Natronomonas kamekura | 用于高分子材料、化学品和燃料的生物制造 | [ | |
| 解脂耶氏酵母 Yarrowia lipolytica | 适合用于油脂及其衍生物的生物合成 | [ | |
| 蓝藻 | 与上转换纳米粒子一起封装,能够创建工程化的微氧细胞工厂 | [ | |
| Minicell | 无染色体细胞,药物递送的优良载体 | [ | |
| Simcell | 无染色体细胞,安全可编程的通用平台 | [ | |
| 复杂基因逻辑回路设计Complex gene logic circuit design | AND、NOT联用型遗传电路设计 | 利用AND和NOT联用的基因电路,巧妙实现了细胞生长过渡到生物合成时间点的精确把控,减少外源电路引入对宿主菌株的代谢负担 | [ |
| CASwitch | 将CRISPR-Cas内环核酸酶与Tet-On3G诱导基因系统相结合而设计的高性能基因电路,用于哺乳动物系统中高性能诱导表达 | [ | |

Fig. 3 Principle of biosensor based on the responses of different components Upon detecting external biomolecular signals encompassing gene, antibody, and enzyme molecules, cells navigate through biosensors meticulously designed with elements such as promoters(a), riboswitch(b), and transcription regulators(c). These biosensors efficiently convert the sensed signals into diverse outputs, including fluorescence signals, electrical signals, and chemical signals. The translated signals are then captured and analyzed outside the cell, providing valuable insights into the intricate cellular responses to external stimuli
| 回路类型 Loop type | 电路原理图 Electrical circuit diagram | 原理 Principle | 来源 Reference |
|---|---|---|---|
| OR | 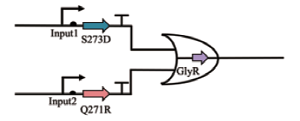 | Input1和Input2分别控制S273和Q271R的形成,二者均可作为甘氨酸合成的前体 | [ |
| NOT | 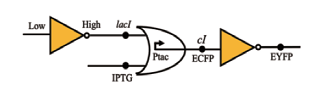 | Lac I和IPTG共同作用于Ptac启动子,作为OR门电路的两个输入。当有高电平输出信号时,会刺激cI和ECFP的表达,进而通过非门抑制EYFP的表达 | [ |
| AND | 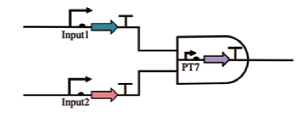 | Input1控制T7 RNA聚合酶基因的转录,Input2控制琥珀酸抑制剂t RNA sup D。当两种成分转录后,合成T7 RNA聚合酶,实现后续的基因表达 | [ |
| NAND | 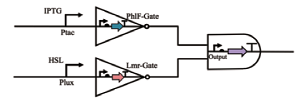 | Phlf和Lmr是两个非门,当IPTG和HSL同时诱导时,黄色荧光蛋白的表达会停止,其余情况正常 | [ |
| NOR | 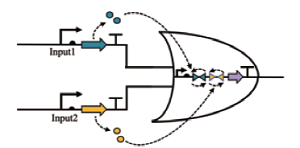 | Input1和Input2合成信号分子,翻转两个终止子的状态,当两个输入信号都表达时,关闭输出启动子 | [ |
Table 2 Summary of basic logic circuit construction
| 回路类型 Loop type | 电路原理图 Electrical circuit diagram | 原理 Principle | 来源 Reference |
|---|---|---|---|
| OR |  | Input1和Input2分别控制S273和Q271R的形成,二者均可作为甘氨酸合成的前体 | [ |
| NOT |  | Lac I和IPTG共同作用于Ptac启动子,作为OR门电路的两个输入。当有高电平输出信号时,会刺激cI和ECFP的表达,进而通过非门抑制EYFP的表达 | [ |
| AND |  | Input1控制T7 RNA聚合酶基因的转录,Input2控制琥珀酸抑制剂t RNA sup D。当两种成分转录后,合成T7 RNA聚合酶,实现后续的基因表达 | [ |
| NAND |  | Phlf和Lmr是两个非门,当IPTG和HSL同时诱导时,黄色荧光蛋白的表达会停止,其余情况正常 | [ |
| NOR |  | Input1和Input2合成信号分子,翻转两个终止子的状态,当两个输入信号都表达时,关闭输出启动子 | [ |
| [1] | Majidian P, Tabatabaei M, Zeinolabedini M, et al. Metabolic engineering of microorganisms for biofuel production[J]. Renew Sustain Energy Rev, 2018, 82: 3863-3885. |
| [2] | Gu PF, Liu LW, Ma QQ, et al. Metabolic engineering of Escherichia coli for the production of isobutanol: a review[J]. World J Microbiol Biotechnol, 2021, 37(10): 168. |
| [3] |
Fidock DA. Eliminating malaria[J]. Science, 2013, 340(6140): 1531-1533.
doi: 10.1126/science.1240539 pmid: 23812705 |
| [4] |
Kannan K, Gibson DG. Yeast genome, by design[J]. Science, 2017, 355(6329): 1024-1025.
doi: 10.1126/science.aam9739 pmid: 28280169 |
| [5] | Joshi YJ, Jawale YK, Athale CA. Modeling the tunability of the dual-feedback genetic oscillator[J]. Phys Rev E, 2020, 101(1-1): 012417. |
| [6] |
Zúñiga A, Guiziou S, Mayonove P, et al. Rational programming of history-dependent logic in cellular populations[J]. Nat Commun, 2020, 11(1): 4758.
doi: 10.1038/s41467-020-18455-z pmid: 32958811 |
| [7] |
Tyler J, Shiu A, Walton J. Revisiting a synthetic intracellular regulatory network that exhibits oscillations[J]. J Math Biol, 2019, 78(7): 2341-2368.
doi: 10.1007/s00285-019-01346-3 pmid: 30929046 |
| [8] | Strelkowa N, Barahona M. Transient dynamics around unstable periodic orbits in the generalized repressilator model[J]. Chaos, 2011, 21(2): 023104. |
| [9] |
Zhang FY, Sun YH, Zhang YH, et al. Independent control of amplitude and period in a synthetic oscillator circuit with modified repressilator[J]. Commun Biol, 2022, 5(1): 23.
doi: 10.1038/s42003-021-02987-1 pmid: 35017621 |
| [10] |
Ozdemir T, Fedorec AJH, Danino T, et al. Synthetic biology and engineered live biotherapeutics: toward increasing system complexity[J]. Cell Syst, 2018, 7(1): 5-16.
doi: S2405-4712(18)30248-5 pmid: 30048620 |
| [11] |
Riglar DT, Giessen TW, Baym M, et al. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation[J]. Nat Biotechnol, 2017, 35: 653-658.
doi: 10.1038/nbt.3879 pmid: 28553941 |
| [12] | Roquet N, Soleimany AP, Ferris AC, et al. Synthetic recombinase-based state machines in living cells[J]. Science, 2016, 353(6297): aad8559. |
| [13] |
Sheth RU, Yim SS, Wu FL, et al. Multiplex recording of cellular events over time on CRISPR biological tape[J]. Science, 2017, 358(6369): 1457-1461.
doi: 10.1126/science.aao0958 pmid: 29170279 |
| [14] |
Adolfsen KJ, Callihan I, Monahan CE, et al. Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering[J]. Nat Commun, 2021, 12(1): 6215.
doi: 10.1038/s41467-021-26524-0 pmid: 34711827 |
| [15] | Zhou PP, Li M, Shen B, et al. Directed coevolution of β-carotene ketolase and hydroxylase and its application in temperature-regulated biosynthesis of astaxanthin[J]. J Agric Food Chem, 2019, 67(4): 1072-1080. |
| [16] | 马平英, 罗雯, 詹怡昕, 等. 枯草芽孢杆菌表达系统研究进展[J]. 江西科学, 2020, 38(6): 867-871. |
| Ma PY, Luo W, Zhan YX, et al. Research progress of Bacillus subtilis expression system[J]. Jiangxi Sci, 2020, 38(6): 867-871. | |
| [17] |
Liu CZ, Qin Y, Li XJ, et al. Preparation and characterization of starch nanoparticles via self-assembly at moderate temperature[J]. Int J Biol Macromol, 2016, 84: 354-360.
doi: 10.1016/j.ijbiomac.2015.12.040 pmid: 26708434 |
| [18] | Dong JW, Li XJ, Yang C, et al. The antioxidant activity and total phenolic and total flavonoid contents of Pyracantha fortuneana fruit can be improved by solid-state fermentation with Rhizopus oryzae and Penicillium commune[J]. Journal of Chinese Pharmaceutical Sciences, 2022, 31(6):452-460. |
| [19] | 晋彪, 张静, 洪坤强, 等. 嗜盐单胞菌利用乙酸盐合成PHB的研究[J]. 化学工业与工程, 2022, 39(5): 119-126. |
| Jin B, Zhang J, Hong KQ, et al. Studies on PHB production in Ha-lomonas using acetate as substrate[J]. Chem Ind Eng, 2022, 39(5): 119-126. | |
| [20] | 赵禹, 刘士琦, 李建, 等. 解脂耶氏酵母作为微生物细胞工厂的应用研究进展[J]. 食品科学, 2021, 42(19): 388-400. |
| Zhao Y, Liu SQ, Li J, et al. Advances in the application of Yarrowia lipolytica as a microbial cell factory[J]. Food Sci, 2021, 42(19): 388-400. | |
| [21] |
Wang WL, Zheng HZ, Jiang J, et al. Engineering micro oxygen factories to slow tumour progression via hyperoxic microenvironments[J]. Nat Commun, 2022, 13(1): 4495.
doi: 10.1038/s41467-022-32066-w pmid: 35918337 |
| [22] | Chen JX, Steel H, Wu YH, et al. Development of aspirin-inducible biosensors in Escherichia coli and SimCells[J]. Appl Environ Microbiol, 2019, 85(6): e02959-e02918. |
| [23] |
Fan C, Davison PA, Habgood R, et al. Chromosome-free bacterial cells are safe and programmable platforms for synthetic biology[J]. Proc Natl Acad Sci USA, 2020, 117(12): 6752-6761.
doi: 10.1073/pnas.1918859117 pmid: 32144140 |
| [24] |
Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression[J]. Nat Biotechnol, 2009, 27(10): 946-950.
doi: 10.1038/nbt.1568 pmid: 19801975 |
| [25] |
De Carluccio G, Fusco V, di Bernardo D. Engineering a synthetic gene circuit for high-performance inducible expression in mammalian systems[J]. Nat Commun, 2024, 15(1): 3311.
doi: 10.1038/s41467-024-47592-y pmid: 38632224 |
| [26] | Schuster SC, Swanson RV, Alex LA, et al. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance[J]. Nature, 1993, 365(6444): 343-347. |
| [27] |
Thouand G, Horry H, Durand MJ, et al. Development of a biosensor for on-line detection of tributyltin with a recombinant bioluminescent Escherichia coli strain[J]. Appl Microbiol Biotechnol, 2003, 62(2-3): 218-225.
pmid: 12883867 |
| [28] | Misawa N, Osaki T, Takeuchi S. Membrane protein-based biosensors[J]. J R Soc Interface, 2018, 15(141): 20170952. |
| [29] |
Maskow T, Kemp R, Buchholz F, et al. What heat is telling us about microbial conversions in nature and technology: from chip- to megacalorimetry[J]. Microb Biotechnol, 2010, 3(3): 269-284.
doi: 10.1111/j.1751-7915.2009.00121.x pmid: 21255327 |
| [30] |
Zhu C, Gerald RE II, Huang J. Micromachined optical fiber sensors for biomedical applications[J]. Methods Mol Biol, 2022, 2393: 367-414.
doi: 10.1007/978-1-0716-1803-5_20 pmid: 34837190 |
| [31] |
Koveal D, Rosen PC, Meyer DJ, et al. A high-throughput multiparameter screen for accelerated development and optimization of soluble genetically encoded fluorescent biosensors[J]. Nat Commun, 2022, 13(1): 2919.
doi: 10.1038/s41467-022-30685-x pmid: 35614105 |
| [32] |
Bourque K, Pétrin D, Sleno R, et al. Distinct conformational dynamics of three G protein-coupled receptors measured using FlAsH-BRET biosensors[J]. Front Endocrinol, 2017, 8: 61.
doi: 10.3389/fendo.2017.00061 pmid: 28439254 |
| [33] |
Alloush HM, Anderson E, Martin AD, et al. A bioluminescent microbial biosensor for in vitro pretreatment assessment of cytarabine efficacy in leukemia[J]. Clin Chem, 2010, 56(12): 1862-1870.
doi: 10.1373/clinchem.2010.145581 pmid: 20921267 |
| [34] |
Dhyani R, Shankar K, Bhatt A, et al. Homogentisic acid-based whole-cell biosensor for detection of alkaptonuria disease[J]. Anal Chem, 2021, 93(10): 4521-4527.
doi: 10.1021/acs.analchem.0c04914 pmid: 33655752 |
| [35] |
Dabirian Y, Li XW, Chen Y, et al. Expanding the dynamic range of a transcription factor-based biosensor in Saccharomyces cerevisiae[J]. ACS Synth Biol, 2019, 8(9): 1968-1975.
doi: 10.1021/acssynbio.9b00144 pmid: 31373795 |
| [36] |
Serganov A, Nudler E. A decade of riboswitches[J]. Cell, 2013, 152(1-2): 17-24.
doi: 10.1016/j.cell.2012.12.024 pmid: 23332744 |
| [37] |
Ma Q, Zhang QW, Xu QY, et al. Systems metabolic engineering strategies for the production of amino acids[J]. Synth Syst Biotechnol, 2017, 2(2): 87-96.
doi: 10.1016/j.synbio.2017.07.003 pmid: 29062965 |
| [38] |
Friedland AE, Lu TK, Wang X, et al. Synthetic gene networks that count[J]. Science, 2009, 324(5931): 1199-1202.
doi: 10.1126/science.1172005 pmid: 19478183 |
| [39] |
Rubens JR, Selvaggio G, Lu TK. Synthetic mixed-signal computation in living cells[J]. Nat Commun, 2016, 7: 11658.
doi: 10.1038/ncomms11658 pmid: 27255669 |
| [40] |
Kong WT, Blanchard AE, Liao C, et al. Engineering robust and tunable spatial structures with synthetic gene circuits[J]. Nucleic Acids Res, 2017, 45(2): 1005-1014.
doi: 10.1093/nar/gkw1045 pmid: 27899571 |
| [41] | Paddon CJ, Westfall PJ, Pitera DJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| [42] |
MacDiarmid JA, Mugridge NB, Weiss JC, et al. Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics[J]. Cancer Cell, 2007, 11(5): 431-445.
pmid: 17482133 |
| [43] |
Ali MK, Liu Q, Liang K, et al. Bacteria-derived minicells for cancer therapy[J]. Cancer Lett, 2020, 491: 11-21.
doi: S0304-3835(20)30379-7 pmid: 32721550 |
| [44] |
Rampley CPN, Davison PA, Qian P, et al. Development of SimCells as a novel chassis for functional biosensors[J]. Sci Rep, 2017, 7(1): 7261.
doi: 10.1038/s41598-017-07391-6 pmid: 28775370 |
| [45] |
Brophy JAN, Voigt CA. Principles of genetic circuit design[J]. Nat Methods, 2014, 11(5): 508-520.
doi: 10.1038/nmeth.2926 pmid: 24781324 |
| [46] | Han L, Shan Q. Pair of residue substitutions at the outer mouth of the channel pore act as inputs for a Boolean logic “OR” gate based on the Glycine receptor[J]. ACS Chem Neurosci, 2020, 11(20): 3409-3417. |
| [47] | Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit[J]. Proc Natl Acad Sci U S A, 2002, 99(26): 16587-16591. |
| [48] |
Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate[J]. Mol Syst Biol, 2007, 3: 133.
pmid: 17700541 |
| [49] |
Stanton BC, Nielsen AAK, Tamsir A, et al. Genomic mining of prokaryotic repressors for orthogonal logic gates[J]. Nat Chem Biol, 2014, 10(2): 99-105.
doi: 10.1038/nchembio.1411 pmid: 24316737 |
| [50] |
Bonnet J, Yin P, Ortiz ME, et al. Amplifying genetic logic gates[J]. Science, 2013, 340(6132): 599-603.
doi: 10.1126/science.1232758 pmid: 23539178 |
| [51] |
Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells[J]. Nat Biotechnol, 2013, 31(5): 448-452.
doi: 10.1038/nbt.2510 pmid: 23396014 |
| [52] | Li XM, Jiang W, Qi QS, et al. A gene circuit combining the endogenous I-E type CRISPR-cas system and a light sensor to produce poly-β-hydroxybutyric acid efficiently[J]. Biosensors, 2022, 12(8): 642. |
| [53] |
Mutalik VK, Guimaraes JC, Cambray G, et al. Precise and reliable gene expression via standard transcription and translation initiation elements[J]. Nat Methods, 2013, 10(4): 354-360.
doi: 10.1038/nmeth.2404 pmid: 23474465 |
| [54] |
Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology[J]. Nat Rev Microbiol, 2014, 12(5): 381-390.
doi: 10.1038/nrmicro3239 pmid: 24686414 |
| [55] | MacKenzie A. From validating to verifying: public appeals in synthetic biology[J]. Sci Cult, 2013, 22(4): 476-496. |
| [1] | CHENG Ting, YUAN Shuai, ZHANG Xiao-yuan, LIN Liang-cai, LI Xin, ZHANG Cui-ying. Research Progress in the Regulation of Isobutanol Synthesis Pathway in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(7): 80-90. |
| [2] | WANG Xiao-mei, YANG Xiao-wei, LI Hui-shang, HE Wei, XIN Zhu-lin. Development Status of Synthetic Biology in Globe and Its Enlightenment [J]. Biotechnology Bulletin, 2023, 39(2): 292-302. |
| [3] | CHEN Xiao-lin, LIU Yang-er, XU Wen-tao, GUO Ming-zhang, LIU Hui-lin. Application of Synthetic Biology Based Whole-cell Biosensor Technology in the Rapid Detection of Food Safety [J]. Biotechnology Bulletin, 2023, 39(1): 137-149. |
| [4] | ZHOU Lin, LIANG Xuan-ming, ZHAO Lei. Biosynthesis of Natural Carotenoids:Progress and Perspective [J]. Biotechnology Bulletin, 2022, 38(7): 119-127. |
| [5] | GUO Xiao-zhen, ZHANG Xue-fu. Analysis of the Development Trend in the Field of Plant Synthetic Biology [J]. Biotechnology Bulletin, 2022, 38(2): 289-296. |
| [6] | ZHAO Yu-xue, WANG Yun, YU Lu-yao, LIU Jing-jing, SI Jin-ping, ZHANG Xin-feng, ZHANG Lei. Structure and Application of C-glycosyltransferases in Plants [J]. Biotechnology Bulletin, 2022, 38(10): 18-28. |
| [7] | YE Min, GAO Jiao-qi, ZHOU Yong-jin. Engineering Non-conventional Yeast Cell Factory for the Biosynthesis of Natural Products [J]. Biotechnology Bulletin, 2021, 37(8): 12-24. |
| [8] | ZHANG Chan, YAO Guang-long, ZHANG Jun-feng, YU Jing, YANG Dong-mei, CHEN Ping, WU You-gen. Research Progress on Patchoulol Molecular Regulation and Synthetic Biology in Pogostemon cablin [J]. Biotechnology Bulletin, 2021, 37(8): 55-64. |
| [9] | YE Jian-wen, CHEN Jiang-nan, ZHANG Xu, Wu Fu-qing, CHEN Guo-qiang. Dynamic Control:An Efficient Strategy for Metabolically Engineering Microbial Cell Factories [J]. Biotechnology Bulletin, 2020, 36(6): 1-12. |
| [10] | CHANG Han-wen, ZHENG Xin-ling, LUO Jian-mei, WANG Min, SHEN Yan-bing. Tolerance Elements and Their Application Progress on the Construction of Highly-efficient Microbial Cell Factory [J]. Biotechnology Bulletin, 2020, 36(6): 13-34. |
| [11] | ZHáNG Hui, TIáN Fáng-fáng, WU Yi. Synthetic Yeást Genome SCRáMbLE [J]. Biotechnology Bulletin, 2020, 36(4): 13-18. |
| [12] | CáO Yán-ting, LIU Yán-feng, LI Jiáng-huá, LIU Long, DU Guo-cheng. ádvánces of Improving the Efficiency of Chemicál Biosynthesis Básed on Cell Subpopulátion Regulátion [J]. Biotechnology Bulletin, 2020, 36(4): 19-25. |
| [13] | LI Jia-xiu, CAI Qian-ru, WU Jie-qun. Research Progresses on the Synthetic Biology of Terpenes in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2020, 36(12): 199-207. |
| [14] | LIU Xin-ping, TAN Yu-meng, ZHANG Xue, FENG Yan, YANG Guang-yu. Biosynthesis of Ganglioside Oligosaccharide Fluoride in Escherichia coli [J]. Biotechnology Bulletin, 2019, 35(8): 162-169. |
| [15] | LIU Yang-er, GUO Ming-zhang, DU Ruo-xi, HE Xiao-yun, HUANG Kun-lun, XU Wen-tao. Advances and Prospects of Synthetic Biology in Lactic Acid Bacteria [J]. Biotechnology Bulletin, 2019, 35(8): 193-204. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||