








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (4): 278-288.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0744
JI Ying-tong( ), GAO Jia-ning, QIAN Meng-ying, PAN Ning, ZHANG Jun, CUI Hong, YAN Xiao-xiao(
), GAO Jia-ning, QIAN Meng-ying, PAN Ning, ZHANG Jun, CUI Hong, YAN Xiao-xiao( )
)
Received:2024-08-02
Online:2025-04-26
Published:2025-04-25
Contact:
YAN Xiao-xiao
E-mail:13596329880@163.com;xiaoxyan@henau.edu.cn
JI Ying-tong, GAO Jia-ning, QIAN Meng-ying, PAN Ning, ZHANG Jun, CUI Hong, YAN Xiao-xiao. Function Study of NtMYC2 Gene on Tobacco Trichome Development[J]. Biotechnology Bulletin, 2025, 41(4): 278-288.
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 用途 Usage |
|---|---|---|---|
| CLNtMYC2a | TCCACTCTCATTCCTCTCACTCA | TTAGCGTGTTTCAGCAACTCTG | NtMYC2克隆 NtMYC2 cloning |
| CLNtMYC2b | TGTCCACTCTCATTCCTCTCACTC | TTAGCGTGTTTCAGCAACTCTG | |
| CLNtMYC2 | ATGACTGATTACAGCTTACCCACC | TTAGCGTGTTTCAGCAACTCTG | NtMYC2a和NtMYC2b 开放阅读框克隆 NtMYC2a and NtMYC2b Open reading frame cloning |
| L25 | CCCCTCACCACAGAGTCTGC | AAGGGTGTTGTTGTCCTCAATCTT | RT-qPCR |
| qRT-NtMYC2a | ATGACTGATTACAGCTTACCCACC | GACAGTAGAAGCACAAGAAGACGG | |
| qRT-NtMYC2b | CTAGTGGTACTACCGATGACAACG | ACAGTAGAGGCACAAGAAGACGG | |
| pC2300-NtMYC2a/2b | GGGGTACCATGACTGATTACAGCTTACCCACC | CGGGATCCGCGTGTTTCAGCAACTCTGG | NtMYC2亚细胞定位载体构建 Vector construction of NtMYC2 subcellular localization |
| MT-NtMYC2a | AGCTTCTGGTGCGATGAAGTC | ATCTTGCTTCCAGTATGGCTAGAC | NtMYC2-KO株系检测 Detection of NtMYC2 knockout lines |
| MT-NtMYC2b | CAGGGGAGGAGAATAAGAACAAG | TCCAGTATGGCTAGACATCTTGTGA | |
| BD-NtJAZ11 | CGGAATTCATGGAGAGAGATTTTATGGG | CGGGATCCTCACCTTGTGTCTACATTATTTTG | NtJAZ11-BD载体构建 Vector construction of NtJAZ11-BD |
| AD-NtMYC2a | TCCCCCGGGATGACTGATTACAGCTTACCCACC | CGGGATCCTTAGCGTGTTTCAGCAACTCTG | NtMYC2a-AD载体构建 Vector construction of NtMYC2a-AD |
| AD-NtMYC2b | GGAATTCCATATGATGACTGATTACAGCTTACCCACC | CGGGATCCTTAGCGTGTTTCAGCAACTCTG | NtMYC2b-AD载体构建 Vector construction of NtMYC2b-AD |
Table 1 Primer sequences
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 用途 Usage |
|---|---|---|---|
| CLNtMYC2a | TCCACTCTCATTCCTCTCACTCA | TTAGCGTGTTTCAGCAACTCTG | NtMYC2克隆 NtMYC2 cloning |
| CLNtMYC2b | TGTCCACTCTCATTCCTCTCACTC | TTAGCGTGTTTCAGCAACTCTG | |
| CLNtMYC2 | ATGACTGATTACAGCTTACCCACC | TTAGCGTGTTTCAGCAACTCTG | NtMYC2a和NtMYC2b 开放阅读框克隆 NtMYC2a and NtMYC2b Open reading frame cloning |
| L25 | CCCCTCACCACAGAGTCTGC | AAGGGTGTTGTTGTCCTCAATCTT | RT-qPCR |
| qRT-NtMYC2a | ATGACTGATTACAGCTTACCCACC | GACAGTAGAAGCACAAGAAGACGG | |
| qRT-NtMYC2b | CTAGTGGTACTACCGATGACAACG | ACAGTAGAGGCACAAGAAGACGG | |
| pC2300-NtMYC2a/2b | GGGGTACCATGACTGATTACAGCTTACCCACC | CGGGATCCGCGTGTTTCAGCAACTCTGG | NtMYC2亚细胞定位载体构建 Vector construction of NtMYC2 subcellular localization |
| MT-NtMYC2a | AGCTTCTGGTGCGATGAAGTC | ATCTTGCTTCCAGTATGGCTAGAC | NtMYC2-KO株系检测 Detection of NtMYC2 knockout lines |
| MT-NtMYC2b | CAGGGGAGGAGAATAAGAACAAG | TCCAGTATGGCTAGACATCTTGTGA | |
| BD-NtJAZ11 | CGGAATTCATGGAGAGAGATTTTATGGG | CGGGATCCTCACCTTGTGTCTACATTATTTTG | NtJAZ11-BD载体构建 Vector construction of NtJAZ11-BD |
| AD-NtMYC2a | TCCCCCGGGATGACTGATTACAGCTTACCCACC | CGGGATCCTTAGCGTGTTTCAGCAACTCTG | NtMYC2a-AD载体构建 Vector construction of NtMYC2a-AD |
| AD-NtMYC2b | GGAATTCCATATGATGACTGATTACAGCTTACCCACC | CGGGATCCTTAGCGTGTTTCAGCAACTCTG | NtMYC2b-AD载体构建 Vector construction of NtMYC2b-AD |

Fig. 1 NtMYC2 gene clone and bioinformatics analysisA: PCR electrophoresis of NtMYC2 gene. B: The phylogenetic tree of NtMYC2 and homologous proteins. C: Multiple sequence alignment of NtMYC2 and homologous proteins of related species; Nt: Nicotiana tabacum; Nsyl: Nicotiana sylvestris; Ntom: Nicotianatomentosiformis; Nb: Nicotiana benthamiana; St: Solanum tuberosum; Sl: Solanum lycopersicum; Ca: Capsicum annuum; Aa: Artemisia annua; At: Arabidopsis thaliana. The black boxes refer to JID domain, TAD motif and bHLH domain

Fig. 2 Analysis of the expression patterns in different tissues of NtMYC2Lower case letters indicate significant differences at P<0.05. The same below
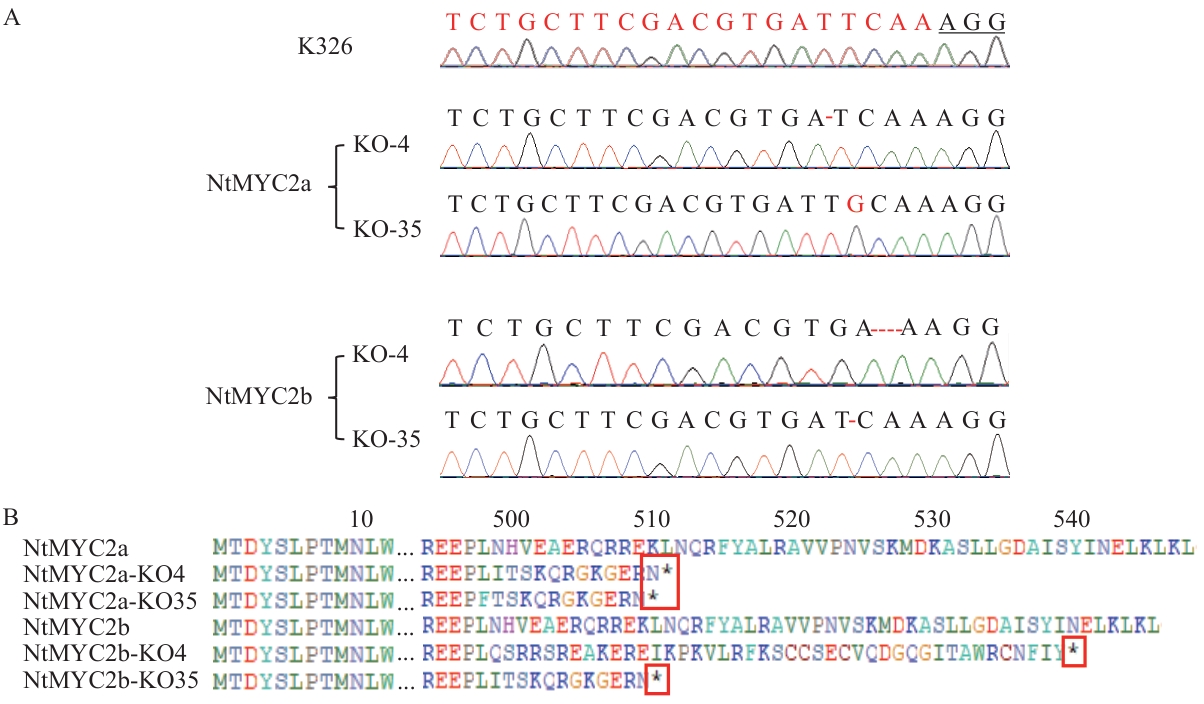
Fig. 5 Identification of NtMYC2 knockout homozygous linesA: Sequencing analysis of sgRNA regions of the homozygous knockout plants. B: Protein translation prediction of NtMYC2 homozygous knockout plants. The red boxes indicate the early termination of translation
| 1 | 崔迪, 孟丽君, 葛秀丽, 等. 铁皮石斛DobHLH51基因克隆及表达特性分析 [J]. 山东农业科学, 2024, 56(5): 9-18. |
| Cui D, Meng LJ, Ge XL, et al. Cloning and expression analysis of DobHLH51 from Dendrobium officinale Kimura et Migo [J]. Shandong Agric Sci, 2024, 56(5): 9-18. | |
| 2 | 寇呈熹, 佟金泉, 马瑞, 等. 藜芦bHLH转录因子分析鉴定 [J]. 中草药, 2022, 53(14): 4476-4485. |
| Kou CX, Tong JQ, Ma R, et al. Analysis of Veratrum nigrum bHLH transcription factor based on transcriptome [J]. Chin Tradit Herb Drugs, 2022, 53(14): 4476-4485. | |
| 3 | Zhao MZ, Morohashi K, Hatlestad G, et al. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci [J]. Development, 2008, 135(11): 1991-1999. |
| 4 | 韩瑶. 基因差异表达与不同倍性小麦根毛长度变异的关系研究 [D]. 北京: 中国农业大学, 2015. |
| Han Y. Study on the relationship between gene differential expression and root hair length variation of wheat with different ploidy [D]. Beijing: China Agricultural University, 2015. | |
| 5 | 王斌, 袁晓, 蒋园园, 等. bHLH96的克隆及其在薄荷萜烯生物合成调控中的功能 [J]. 生物技术通报, 2024, 40(1): 281-293. |
| Wang B, Yuan X, Jiang YY, et al. Cloning of bHLH96 gene and its roles in regulating the biosynthesis of peppermint terpenes [J]. Biotechnol Bull, 2024, 40(1): 281-293. | |
| 6 | Outchkourov NS, Carollo CA, Gomez-Roldan V, et al. Control of anthocyanin and non-flavonoid compounds by anthocyanin-regulating MYB and bHLH transcription factors in Nicotiana benthamiana leaves [J]. Front Plant Sci, 2014, 5: 519. |
| 7 | Liang YF, Ma F, Li BY, et al. A bHLH transcription factor, SlbHLH96, promotes drought tolerance in tomato [J]. Hortic Res, 2022, 9: uhac198. |
| 8 | Seo JS, Joo J, Kim MJ, et al. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice [J]. Plant J, 2011, 65(6): 907-921. |
| 9 | Wang YG, Wang S, Tian Y, et al. Functional characterization of a sugar beet BvbHLH93 transcription factor in salt stress tolerance [J]. Int J Mol Sci, 2021, 22(7): 3669. |
| 10 | Geng JJ, Liu JH. The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene [J]. J Exp Bot, 2018, 69(10): 2677-2692. |
| 11 | Zhao Q, Xiang XH, Liu D, et al. Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis [J]. Front Plant Sci, 2018, 9: 381. |
| 12 | 高丽华, 刘博欣, 李金博, 等. 陆地棉bHLH转录因子GhMYC4基因的克隆及功能分析 [J]. 中国农业科技导报, 2016, 18(5): 33-41. |
| Gao LH, Liu BX, Li JB, et al. Cloning and function analysis of b HLH transcription factor gene GhMYC4 from Gossypium hirsutism L [J]. J Agric Sci Technol, 2016, 18(5): 33-41. | |
| 13 | Kazan K, Manners JM. MYC2: the master in action [J]. Mol Plant, 2013, 6(3): 686-703. |
| 14 | Gao CH, Qi SH, Liu KG, et al. MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis [J]. Plant Physiol Biochem, 2016, 108: 63-70. |
| 15 | Niu YJ, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis [J]. J Exp Bot, 2011, 62(6): 2143-2154. |
| 16 | Çevik V, Kidd BN, Zhang PJ, et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis [J]. Plant Physiol, 2012, 160(1): 541-555. |
| 17 | Abe H, Urao T, Ito T, et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling [J]. Plant Cell, 2003, 15(1): 63-78. |
| 18 | Wei M, Liu QG, Wang ZC, et al. PuHox52-mediated hierarchical multilayered gene regulatory network promotes adventitious root formation in Populus ussuriensis [J]. New Phytol, 2020, 228(4): 1369-1385. |
| 19 | Zhang HT, Hedhili S, Montiel G, et al. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus [J]. Plant J, 2011, 67(1): 61-71. |
| 20 | Zhang HB, Bokowiec MT, Rushton PJ, et al. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis [J]. Mol Plant, 2012, 5(1): 73-84. |
| 21 | 悦曼芳, 张春, 郑登俞, 等. 玉米转录因子ZmbHLH91对非生物逆境胁迫的应答 [J]. 作物学报, 2022, 48(12): 3004-3017. |
| Yue MF, Zhang C, Zheng DY, et al. Response of maize transcriptional factor ZmbHLH91 to abiotic stress [J]. Acta Agron Sin, 2022, 48(12): 3004-3017. | |
| 22 | Fu JY, Wang LP, Pei WZ, et al. ZmEREB92 interacts with ZmMYC2 to activate maize terpenoid phytoalexin biosynthesis upon Fusarium graminearum infection through jasmonic acid/ethylene signaling [J]. New Phytol, 2023, 237(4): 1302-1319. |
| 23 | 袁铭远. AabHLH113整合JA和ABA信号调控青蒿素生物合成的功能研究 [D]. 重庆: 西南大学, 2023. |
| Yuan MY. Study on the function of AabHLH113 integrating JA and ABA signals to regulate artemisinin biosynthesis [D]. Chongqing: Southwest University, 2023. | |
| 24 | Hua B, Chang J, Wu ML, et al. Mediation of JA signalling in glandular trichomes by the woolly/SlMYC1 regulatory module improves pest resistance in tomato [J]. Plant Biotechnol J, 2021, 19(2): 375-393. |
| 25 | Xu JS, van Herwijnen ZO, Dräger DB, et al. SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells [J]. Plant Cell, 2018, 30(12): 2988-3005. |
| 26 | 余兰, 王浩然, 张莹, 等. 转录因子MYCs调控番茄表皮毛萜类化合物的分子机制研究进展 [J]. 中国农学通报, 2022, 38(6): 87-93. |
| Yu L, Wang HR, Zhang Y, et al. Transcription factor MYCs regulating terpenoids in tomato trichomes: research progress on molecular mechanism [J]. Chin Agric Sci Bull, 2022, 38(6): 87-93. | |
| 27 | Feng ZX, Sun L, Dong MM, et al. Identification and functional characterization of CsMYCs in cucumber glandular trichome development [J]. Int J Mol Sci, 2023, 24(7): 6435. |
| 28 | Yuan MY, Sheng YG, Bao JJ, et al. AaMYC3 bridges the regulation of glandular trichome density and artemisinin biosynthesis in Artemisia annua [J]. Plant Biotechnol J, 2024. |
| 29 | Guan YQ, Jiang L, Wang Y, et al. CmMYC2-CmMYBML1 module orchestrates the resistance to herbivory by synchronously regulating the trichome development and constitutive terpene biosynthesis in Chrysanthemum [J]. New Phytol, 2024, 244(3): 914-933. |
| 30 | 孟盈, 闫筱筱, 王召军, 等. NtCycB2基因表达对烟草腺毛发生的影响 [J]. 中国烟草学报, 2019, 25(2): 85-92, 98. |
| Meng Y, Yan XX, Wang ZJ, et al. Effects of NtCycB2 expression on tobacco trichomes [J]. Acta Tabacaria Sin, 2019, 25(2): 85-92, 98. | |
| 31 | 潘影, 张振远, 滕环瑜, 等. 烟草腺毛生物反应器材料的创制与性状分析 [J]. 烟草科技, 2023, 56(7): 1-7. |
| Pan Y, Zhang ZY, Teng HY, et al. Creation and characterization of tobacco glandular trichome bioreactor material [J]. Tob Sci Technol, 2023, 56(7): 1-7. | |
| 32 | Qi TC, Song SS, Ren QC, et al. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana [J]. Plant Cell, 2011, 23(5): 1795-1814. |
| 33 | Hu HY, He X, Tu LL, et al. GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like [J]. Plant J, 2016, 88(6): 921-935. |
| 34 | Hua B, Chang J, Xu ZJ, et al. HOMEODOMAIN PROTEIN8 mediates jasmonate-triggered trichome elongation in tomato [J]. New Phytol, 2021, 230(3): 1063-1077. |
| 35 | Yan XX, Cui LP, Liu XY, et al. NbJAZ3 is required for jasmonate-meditated glandular trichome development in Nicotiana benthamiana [J]. Physiol Plant, 2022, 174(2): e13666. |
| 36 | 徐寒池, 徐梦晓, 滕环瑜, 等. 烟草NtCycB2基因通过茉莉酮酸途径调控腺毛发生 [J]. 中国烟草学报, 2023, 29(4): 96-104. |
| Xu HC, Xu MX, Teng HY, et al. Tobacco NtCycB2 regulates glandular trichomes generation via jasmonic acid pathway [J]. Acta Tabacaria Sin, 2023, 29(4): 96-104. | |
| 37 | 张娇. 两个bHLH转录因子(AtLPl和AtLP2)在拟南芥细胞伸长生长中的功能研究 [D]. 武汉: 华中师范大学, 2019. |
| Zhang J. Function of two bHLH transcription factors (AtLPl and AtLP2) in elongation and growth of Arabidopsis thaliana cells [D]. Wuhan: Central China Normal University, 2019. | |
| 38 | Liu ZH, Chen Y, Wang NN, et al. A basic helix-loop-helix protein (GhFP1) promotes fibre elongation of cotton (Gossypium hirsutum) by modulating brassinosteroid biosynthesis and signalling [J]. New Phytol, 2020, 225(6): 2439-2452. |
| 39 | Vaughan Symonds V, Hatlestad G, Lloyd AM. Natural allelic variation defines a role for ATMYC1: trichome cell fate determination [J]. PLoS Genet, 2011, 7(6): e1002069. |
| 40 | Cheng ZW, Sun L, Qi TC, et al. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis [J]. Mol Plant, 2011, 4(2): 279-288. |
| 41 | Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses [J]. Plant Cell, 2011, 23(2): 701-715. |
| 42 | Wang S, Wang YB, Yang R, et al. Genome-wide identification and analysis uncovers the potential role of JAZ and MYC families in potato under abiotic stress [J]. Int J Mol Sci, 2023, 24(7): 6706. |
| 43 | Zhang HY, Li WJ, Niu DX, et al. Tobacco transcription repressors NtJAZ: Potential involvement in abiotic stress response and glandular trichome induction [J]. Plant Physiol Biochem, 2019, 141: 388-397. |
| [1] | LU Yong-jie, XIA Hai-qian, LI Yong-ling, ZHANG Wen-jian, YU Jing, ZHAO Hui-na, WANG Bing, XU Ben-bo, LEI Bo. Cloning and Expression Analysis of AP2/ERF Transcription Factor NtESR2 in Nicotiana tabacum [J]. Biotechnology Bulletin, 2025, 41(4): 266-277. |
| [2] | MA Yao-wu, ZHANG Qi-yu, YANG Miao, JIANG Cheng, ZHANG Zhen-yu, ZHANG Yi-lin, LI Meng-sha, XU Jia-yang, ZHANG Bin, CUI Guang-zhou, JIANG Ying. Screening, Indentification and Promotion Performance Investigation of Tobacco Growth-promoting Rhizobacteria [J]. Biotechnology Bulletin, 2025, 41(3): 271-281. |
| [3] | HAN Meng-qiao, WU Jiang, LI Li-hua, WANG Zhao-yi, DENG Xi, WEI Feng-jie, REN Min, SUN Yang-yang, LI Fu-xin. Establishment and Optimization of a Tobacco Chromosome Preparation System [J]. Biotechnology Bulletin, 2025, 41(3): 44-50. |
| [4] | XIANG Bo-ka, ZHOU Zuan-zuan, FENG Jia-hui, XIA Chen, LI Qi, CHEN Chun. Isolation and Identification of a Fungus from Moldy Tobacco Leaf and Study on Its Mold-causing Factors [J]. Biotechnology Bulletin, 2025, 41(2): 321-330. |
| [5] | ZHANG Man-yu, DONG Jia-cheng, GOU Fu-fan, GONG Chao-hui, LIU Qian, SUN Wen-liang, KONG zhen, HAO Jie, WANG Min, TIAN Chao-guang. Cloning, Expression, Characterization and Application of the Pectin Esterase MtCE12-1 from Myceliophthora thermophila [J]. Biotechnology Bulletin, 2024, 40(9): 291-300. |
| [6] | LIAO Yang-mei, ZHAO Guo-chun, WENG Xue-huang, JIA Li-ming, CHEN Zhong. Transcriptome Sequencing of Male Sterile Buds at Different Developmental Stages in Sapindus mukorossi ‘Qirui’ [J]. Biotechnology Bulletin, 2024, 40(7): 197-206. |
| [7] | YU Xin-lei, HE Jie-wang, LIN Guo-ping, LI Jin-hai, WANG Da-ai, YUAN Yue-bin, LIU Sheng-gao, LI Zhi-hao, TAO De-xin. Metabolome Difference Analysis of Fermented Cigar Tobacco Leaves in Summer and Winter [J]. Biotechnology Bulletin, 2024, 40(6): 260-270. |
| [8] | WANG Hao-jie, CHANG Dong, LI Jun-ying, MENG Hao-guang, JIANG Shi-jun, ZHOU Shuo-ye, CUI Jiang-kuan. Analysis of Microbial Community Changes and Stress-resistant Enzyme Activities of Flue-cured Tobacco Three-stage Seedling Raising in Different Habitats [J]. Biotechnology Bulletin, 2024, 40(4): 242-254. |
| [9] | LI Can, JIANG Xiang-ning, GAI Ying. Cloning of the LkF3H2 Gene in Larix kaempferi and Its Function in Regulating Flavonoid Metabolism [J]. Biotechnology Bulletin, 2024, 40(2): 245-252. |
| [10] | WU Qiao-yin, SHI You-zhi, LI Lin-lin, PENG Zheng, TAN Zai-yu, LIU Li-ping, ZHANG Juan, PAN Yong. In Situ Screening of Carotenoid Degrading Strains and the Application in Improving Quality and Aroma of Cigar [J]. Biotechnology Bulletin, 2023, 39(9): 192-201. |
| [11] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [12] | LIU Zhen-yin, DUAN Zhi-zhen, PENG Ting, WANG Tong-xin, WANG Jian. Establishment and Optimization of Virus-induced Gene Silencing System in Bougainvillea peruviana ‘Thimma’ [J]. Biotechnology Bulletin, 2023, 39(7): 123-130. |
| [13] | LI Wen-chen, LIU Xin, KANG Yue, LI Wei, QI Ze-zheng, YU Lu, WANG Fang. Optimization and Application of Tobacco Rattle Virus-induced Gene Silencing System in Soybean [J]. Biotechnology Bulletin, 2023, 39(7): 143-150. |
| [14] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [15] | SHEN Yun-xin, SHI Zhu-feng, ZHOU Xu-dong, LI Ming-gang, ZHANG Qing, FENG Lu-yao, CHEN Qi-bin, YANG Pei-wen. Isolation, Identification and Bio-activity of Three Bacillus Strains with Biocontrol Function [J]. Biotechnology Bulletin, 2023, 39(3): 267-277. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||