








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 301-310.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0366
JIANG Tian-wei1( ), LI Ya-jiao2, MA Pei-jie2, CHEN Cai-jun2, LIU Xiao-xia2, CHEN Ying2(
), LI Ya-jiao2, MA Pei-jie2, CHEN Cai-jun2, LIU Xiao-xia2, CHEN Ying2( ), WANG Xiao-li2(
), WANG Xiao-li2( )
)
Received:2025-04-07
Online:2025-11-26
Published:2025-12-09
Contact:
CHEN Ying, WANG Xiao-li
E-mail:JiangTianwei_GIP@163.com;379145306@qq.com;WangXiaoli_GIP@163.com
JIANG Tian-wei, LI Ya-jiao, MA Pei-jie, CHEN Cai-jun, LIU Xiao-xia, CHEN Ying, WANG Xiao-li. Whole-genome DNA Methylation Analysis during the Flowering Processof Medicago truncatula[J]. Biotechnology Bulletin, 2025, 41(11): 301-310.
样本 Sample | 总读取数 Total reads | 比对读取数 Mapped reads | 唯一比对读取数 Uniquely reads | 测序深度 Seqencing depth | 唯一比对率 Uniquely mapped reads (%) | 比对率 Mapping rate (%) | Q30 (%) | 转换率 Conversion rate (%) |
|---|---|---|---|---|---|---|---|---|
| FP_1 | 107573870 | 62923962 | 43437098 | 34.38 | 40.38 | 58.49 | 90.98 | 99.69 |
| FP_2 | 93350734 | 52038408 | 34920690 | 29.86 | 37.41 | 55.75 | 91.34 | 99.67 |
| FP_3 | 114432912 | 66064890 | 45284390 | 36.65 | 39.57 | 57.73 | 91.97 | 99.65 |
| NP_1 | 103921110 | 60296822 | 44356076 | 33.25 | 42.68 | 58.02 | 91.44 | 99.67 |
| NP_2 | 120328788 | 72294208 | 50808854 | 38.52 | 42.23 | 60.08 | 91.87 | 99.59 |
| NP_3 | 129107620 | 74594082 | 51931090 | 41.32 | 40.22 | 57.78 | 91.59 | 99.61 |
Table 1 Processing of whole-genome methylation sequencing data of Medicago truncatula
样本 Sample | 总读取数 Total reads | 比对读取数 Mapped reads | 唯一比对读取数 Uniquely reads | 测序深度 Seqencing depth | 唯一比对率 Uniquely mapped reads (%) | 比对率 Mapping rate (%) | Q30 (%) | 转换率 Conversion rate (%) |
|---|---|---|---|---|---|---|---|---|
| FP_1 | 107573870 | 62923962 | 43437098 | 34.38 | 40.38 | 58.49 | 90.98 | 99.69 |
| FP_2 | 93350734 | 52038408 | 34920690 | 29.86 | 37.41 | 55.75 | 91.34 | 99.67 |
| FP_3 | 114432912 | 66064890 | 45284390 | 36.65 | 39.57 | 57.73 | 91.97 | 99.65 |
| NP_1 | 103921110 | 60296822 | 44356076 | 33.25 | 42.68 | 58.02 | 91.44 | 99.67 |
| NP_2 | 120328788 | 72294208 | 50808854 | 38.52 | 42.23 | 60.08 | 91.87 | 99.59 |
| NP_3 | 129107620 | 74594082 | 51931090 | 41.32 | 40.22 | 57.78 | 91.59 | 99.61 |

Fig. 1 Variations in DNA methylation from the vegetative stage to the flowering stage in M. truncatulaA: Sample correlation analysis. B: Methylation level statistics for the three sequences. C: Methylation distribution in gene body regions and their flanking regions for the three sequences. FP refers to the flowering stage, and NP refers to the vegetative stage. The same below
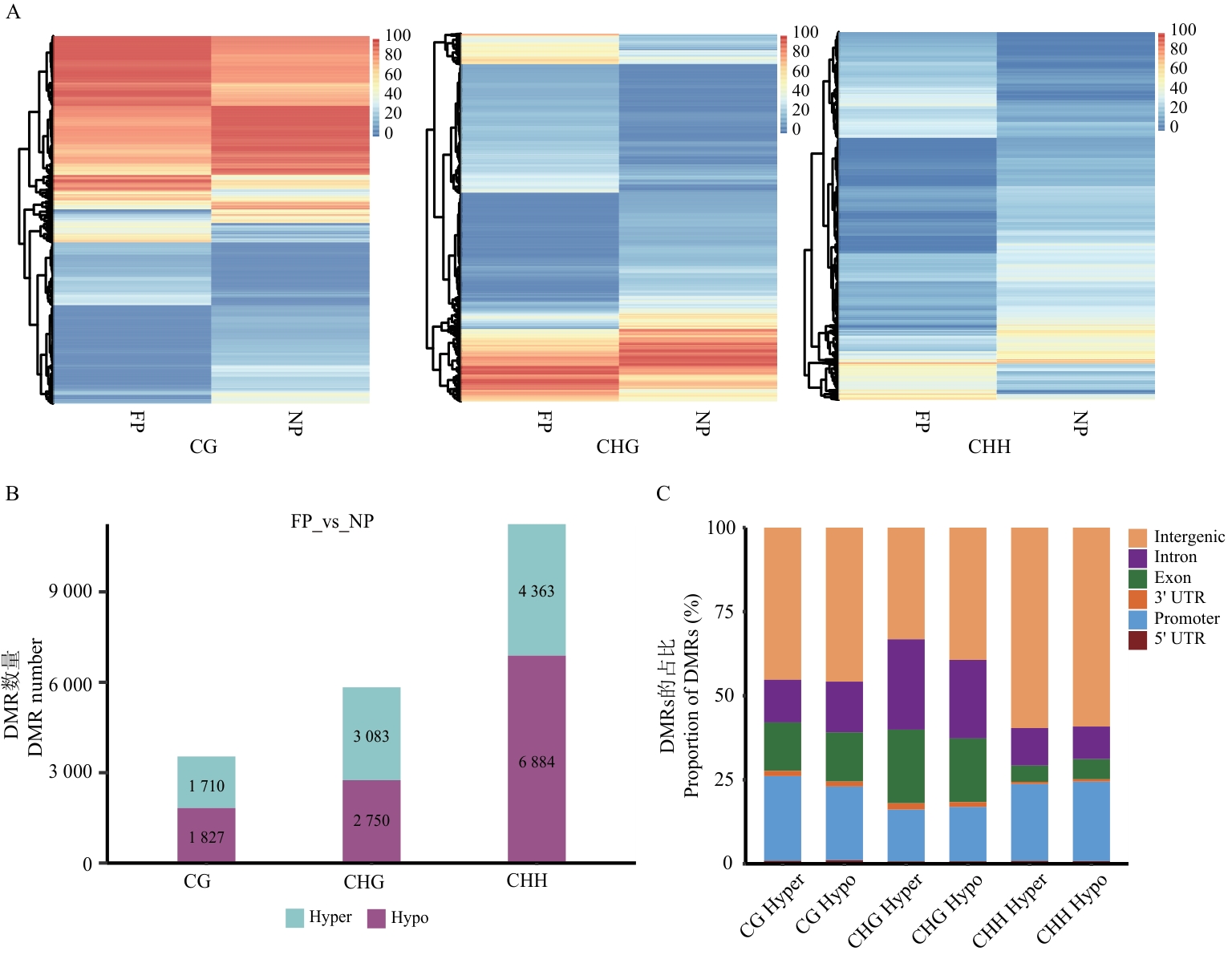
Fig. 2 Differentially methylated region analysis between FP and NPA: Clustering heatmap of differentially methylated regions (DMRs) in the three sequences. B: Statistical chart of the number of DMRs. C: Proportional distribution of DMRs in the genome. In figure B and C, Hyper indicates hypermethylation, and Hypo indicates hypomethylation
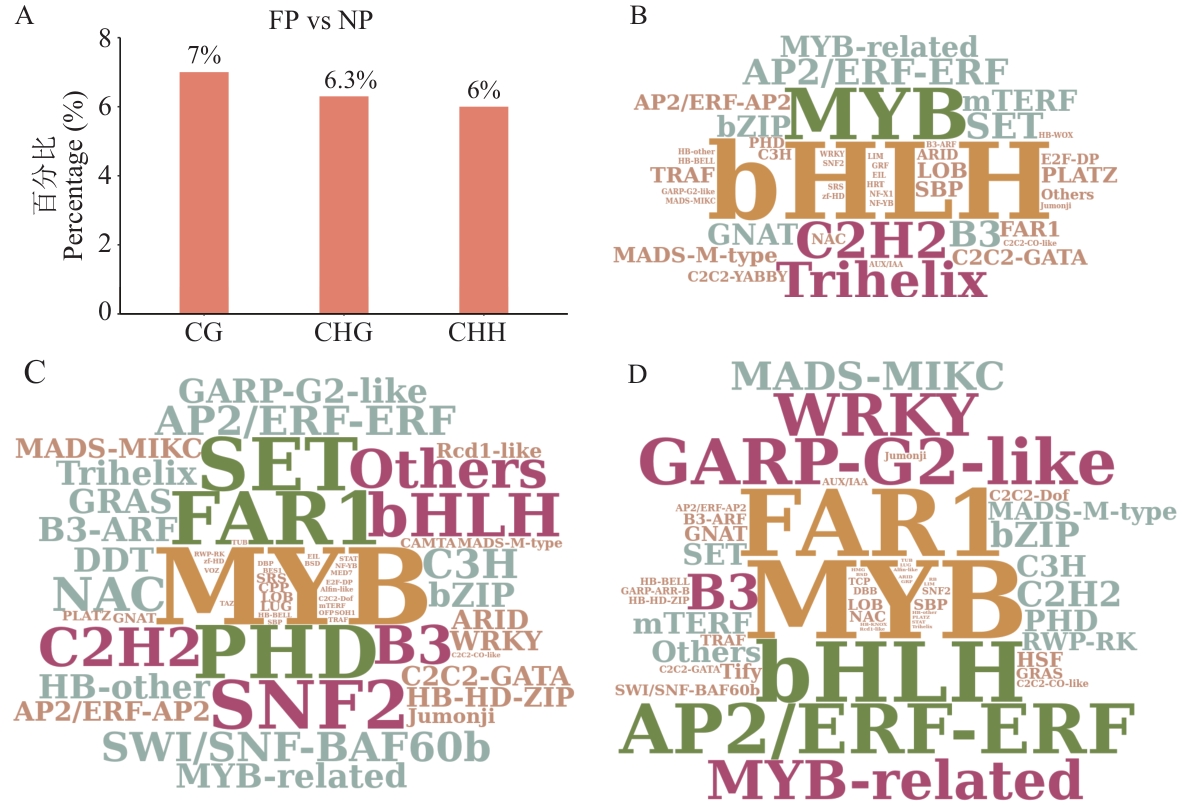
Fig. 4 Transcription factor analysis associated with DMRsA: Proportion of transcription factor-encoding genes among DMR-associated genes in FP vs NP. B, C, D: Word clouds showing the abundance of DMR-associated transcription factor families under three sequence contexts. Figure B corresponds to CG, Figure C to CHG, and Figure D to CHH; the larger the text in Figure B, C, and D, the higher the abundance

Fig. 5 Protein-protein interaction network of transcription factor-encoding genesA: Protein-protein interaction network under CG context. B: Protein-protein interaction network under CHG context. C: Protein-protein interaction network under CHH context. Nodes indicate proteins, edges indicate interaction relationships, and red text next to the nodes indicates the gene IDs of key nodes
序列类型 Type | 基因 Gene | 去甲基化 Hypo-DMR | 超甲基化 Hyper-DMR |
|---|---|---|---|
| CG | ACTR4 | - | Up |
| GA20ox1 | - | Up | |
| CRY1 | - | Up | |
| UBOX13 | - | Up | |
| NFYB3 | - | Up | |
| CRY2 | - | Up | |
| MYB1 | - | Up | |
| CHS | - | Up | |
| SRO1 | Down | - | |
| BHLH130 | Down | - | |
| CHG | CCX5 | - | Up |
| VOZ1 | - | Up | |
| ACTR4 | - | Up | |
| ATX2 | - | Up | |
| FKF1 | - | Up | |
| MYB75 | - | Up | |
| PHYA | - | Up | |
| ELF3 | - | Up | |
| UPF2 | Down | - | |
| ELF6 | Down | - | |
| BRN1 | Down | - | |
| CHS | Down | - | |
| COL 2 | Down | - | |
| CHH | EZA1 | - | Up |
| PEPPER | - | Up | |
| MYB75 | - | Up | |
| CSNK2A | - | Up | |
| CHS9 | - | Up | |
| CHS2 | Down | - | |
| DMR6-LIKE OXYGENASE 2 | Down | - | |
| RENT3 | Down | - | |
| FD | Down | - | |
| CCA1 | Down | - | |
| DMR6-LIKE OXYGENASE 2 | Down | - | |
| bHLH122 | Down | - | |
| MYST family 2 | Down | - | |
| COL 9 | Down | - | |
| EZA1 | Down | - | |
| BRN1 | Down | - | |
| AGL18 | Down | - | |
| FT | Down | - | |
| MYB114 | Down | - | |
| LHY | Down | - | |
| MYB1 | Down | - | |
| CHS | Down | - | |
| ZTL | Down | - | |
| COL2 | Down | - |
Table 2 DMR-associated genes related to photoperiod
序列类型 Type | 基因 Gene | 去甲基化 Hypo-DMR | 超甲基化 Hyper-DMR |
|---|---|---|---|
| CG | ACTR4 | - | Up |
| GA20ox1 | - | Up | |
| CRY1 | - | Up | |
| UBOX13 | - | Up | |
| NFYB3 | - | Up | |
| CRY2 | - | Up | |
| MYB1 | - | Up | |
| CHS | - | Up | |
| SRO1 | Down | - | |
| BHLH130 | Down | - | |
| CHG | CCX5 | - | Up |
| VOZ1 | - | Up | |
| ACTR4 | - | Up | |
| ATX2 | - | Up | |
| FKF1 | - | Up | |
| MYB75 | - | Up | |
| PHYA | - | Up | |
| ELF3 | - | Up | |
| UPF2 | Down | - | |
| ELF6 | Down | - | |
| BRN1 | Down | - | |
| CHS | Down | - | |
| COL 2 | Down | - | |
| CHH | EZA1 | - | Up |
| PEPPER | - | Up | |
| MYB75 | - | Up | |
| CSNK2A | - | Up | |
| CHS9 | - | Up | |
| CHS2 | Down | - | |
| DMR6-LIKE OXYGENASE 2 | Down | - | |
| RENT3 | Down | - | |
| FD | Down | - | |
| CCA1 | Down | - | |
| DMR6-LIKE OXYGENASE 2 | Down | - | |
| bHLH122 | Down | - | |
| MYST family 2 | Down | - | |
| COL 9 | Down | - | |
| EZA1 | Down | - | |
| BRN1 | Down | - | |
| AGL18 | Down | - | |
| FT | Down | - | |
| MYB114 | Down | - | |
| LHY | Down | - | |
| MYB1 | Down | - | |
| CHS | Down | - | |
| ZTL | Down | - | |
| COL2 | Down | - |
| [23] | Taoka KI, Ohki I, Tsuji H, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen [J]. Nature, 2011, 476(7360): 332-335. |
| [24] | Qin ZR, Bai YX, Muhammad S, et al. Divergent roles of FT-like 9 in flowering transition under different day lengths in Brachypodium distachyon [J]. Nat Commun, 2019, 10(1): 812. |
| [25] | Yang ZF, Yan HD, Nie G, et al. Induction of flowering under long-day photoperiod requires DNA hypermethylation in orchardgrass [J]. J Exp Bot, 2025, 76(9): 2557-2572. |
| [26] | Zhou Q, Jia CL, Ma WX, et al. MYB transcription factors in alfalfa (Medicago sativa): genome-wide identification and expression analysis under abiotic stresses [J]. PeerJ, 2019, 7: e7714. |
| [27] | Zhang LC, Liu GX, Jia JZ, et al. The wheat MYB-related transcription factor TaMYB72 promotes flowering in rice [J]. J Integr Plant Biol, 2016, 58(8): 701-704. |
| [28] | Zhu L, Guan YX, Liu YN, et al. Regulation of flowering time in Chrysanthemum by the R2R3 MYB transcription factor CmMYB2 is associated with changes in gibberellin metabolism [J]. Hortic Res, 2020, 7: 96. |
| [29] | Liu YW, Li X, Li KW, et al. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis [J]. PLoS Genet, 2013, 9(10): e1003861. |
| [30] | Aslam M, Jakada BH, Fakher B, et al. Genome-wide study of pineapple (Ananas comosus L.) bHLH transcription factors indicates that cryptochrome-interacting bHLH2 (AcCIB2) participates in flowering time regulation and abiotic stress response [J]. BMC Genomics, 2020, 21(1): 735. |
| [31] | Zhou Q, Cui Y, Dong R, et al. Integrative analyses of transcriptomes and metabolomes reveal associated genes and metabolites with flowering regulation in common vetch (Vicia sativa L.) [J]. Int J Mol Sci, 2022, 23(12): 6818. |
| [32] | Sun H, Guo ZA, Gao LF, et al. DNA methylation pattern of Photoperiod-B1 is associated with photoperiod insensitivity in wheat (Triticum aestivum) [J]. New Phytol, 2014, 204(3): 682-692. |
| [33] | Sun Q, Qiao J, Zhang S, et al. Changes in DNA methylation assessed by genomic bisulfite sequencing suggest a role for DNA methylation in cotton fruiting branch development [J]. PeerJ, 2018, 6: e4945. |
| [34] | Song QX, Zhang TZ, Stelly DM, et al. Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons [J]. Genome Biol, 2017, 18(1): 99. |
| [1] | Qi QY, Hu BC, Jiang WY, et al. Advances in plant epigenome editing research and its application in plants [J]. Int J Mol Sci, 2023, 24(4): 3442. |
| [2] | Qiao S, Song W, Hu WT, et al. The role of plant DNA methylation in development, stress response, and crop breeding [J]. Agronomy (Basel), 2024, 15(1): 94. |
| [3] | Zeng YB, Dawe RK, Gent JI. Natural methylation epialleles correlate with gene expression in maize [J]. Genetics, 2023, 225(2): iyad146. |
| [4] | Mahmood T, He SP, Abdullah M, et al. Epigenetic insight into floral transition and seed development in plants [J]. Plant Sci, 2024, 339: 111926. |
| [5] | Kumar S, Mohapatra T. Dynamics of DNA methylation and its functions in plant growth and development [J]. Front Plant Sci, 2021, 12: 596236. |
| [6] | Sun M. Advances in plant epigenetic regulation of abiotic stress response[J]. Theoretical and Natural Science, 2025, 90: 81-87. |
| [7] | Rehman S, Bahadur S, Xia W. An overview of floral regulatory genes in annual and perennial plants [J]. Gene, 2023, 885: 147699. |
| [8] | Shi ZY, Zhao WQ, Li CR, et al. Overexpression of the Chrysanthemum lavandulifolium ROS1 gene promotes flowering in Arabidopsis thaliana by reducing the methylation level of CONSTANS [J]. Plant Sci, 2024, 342: 112019. |
| [9] | Shi MM, Wang CL, Wang P, et al. Role of methylation in vernalization and photoperiod pathway: a potential flowering regulator? [J]. Hortic Res, 2023, 10(10): uhad174. |
| [10] | Xie HJ, Li XC, Sun YL, et al. DNA methylation of the autonomous pathway is associated with flowering time variations in Arabidopsis thaliana [J]. Int J Mol Sci, 2024, 25(13): 7478. |
| [11] | Sala-Cholewa K, Tomasiak A, Nowak K, et al. DNA methylation analysis of floral parts revealed dynamic changes during the development of homostylous Fagopyrum tataricum and heterostylous F. esculentum flowers [J]. BMC Plant Biol, 2024, 24(1): 448. |
| [12] | He GM, Zhu XP, Elling AA, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids [J]. Plant Cell, 2010, 22(1): 17-33. |
| [35] | Hsieh TF, Ibarra CA, Silva P, et al. Genome-wide demethylation of Arabidopsis endosperm [J]. Science, 2009, 324(5933): 1451-1454. |
| [13] | Xu W, Yang TQ, Dong X, et al. Genomic DNA methylation analyses reveal the distinct profiles in Castor bean seeds with persistent endosperms [J]. Plant Physiol, 2016, 171(2): 1242-1258. |
| [14] | Vagner B, Senjuti S, Yun K, et al. The Medicago Truncatula Genome [M]. Cham, Switzerland : Springer, 2022. |
| [15] | Beck D, Ben Maamar M, Skinner MK. Genome-wide CpG density and DNA methylation analysis method (MeDIP, RRBS, and WGBS) comparisons [J]. Epigenetics, 2022, 17(5): 518-530. |
| [16] | Noh B, Lee SH, Kim HJ, et al. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time [J]. Plant Cell, 2004, 16(10): 2601-2613. |
| [17] | Lin XW, Yuan TT, Guo H, et al. The regulation of chromatin configuration at AGAMOUS locus by LFR-SYD-containing complex is critical for reproductive organ development in Arabidopsis [J]. Plant J, 2023, 116(2): 478-496. |
| [18] | Sachdev S, Biswas R, Roy A, et al. The Arabidopsis arid-hmg dna-binding protein 15 modulates jasmonic acid signaling by regulating MYC2 during pollen development [J]. Plant Physiol, 2024, 196(2): 996-1013. |
| [19] | Bartels A, Han Q, Nair P, et al. Dynamic DNA methylation in plant growth and development [J]. Int J Mol Sci, 2018, 19(7): 2144. |
| [20] | Kondo H, Shiraya T, Wada KC, et al. Induction of flowering by DNA demethylation in Perilla frutescens and Silene armeria: Heritability of 5-azacytidine-induced effects and alteration of the DNA methylation state by photoperiodic conditions [J]. Plant Sci, 2010, 178(3): 321-326. |
| [21] | Yang HX, Chang F, You CJ, et al. Whole-genome DNA methylation patterns and complex associations with gene structure and expression during flower development in Arabidopsis [J]. Plant J, 2015, 81(2): 268-281. |
| [22] | Wu X, Chen SY, Lin F, et al. Comparative and functional analysis unveils the contribution of photoperiod to DNA methylation, sRNA accumulation, and gene expression variations in short-day and long-day grasses [J]. Plant J, 2024, 118(6): 1955-1971. |
| [1] | ZHANG Chao-chao, HAN Kai-yuan, WANG Tong, CHEN Zhong. Cloning and Functional Analysis of PtoYABBY2 and PtoYABBY12 in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(9): 256-264. |
| [2] | SHI Fa-chao, JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian. Cloning and Functional Analysis of LcTFL1 Gene in Litchi chinensis Sonn. [J]. Biotechnology Bulletin, 2025, 41(9): 159-167. |
| [3] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [4] | JIANG Tian-wei, MA Pei-jie, LI Ya-jiao, CHEN Cai-jun, LIU Xiao-xia, WANG Xiao-li. Metabolic Response Analysis of Brachypodium distachyon to Photoperiods [J]. Biotechnology Bulletin, 2025, 41(7): 237-247. |
| [5] | LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 181-192. |
| [6] | GUO Tao, AI Li-jiao, ZOU Shi-hui, ZHOU Ling, LI Xue-mei. Functional Study of CjRAV1 from Camellia japonica in Regulating Flowering Delay [J]. Biotechnology Bulletin, 2025, 41(6): 208-217. |
| [7] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [8] | HU Ruo-qun, ZENG Jing-jing, LIANG Wan-feng, CAO Jia-yu, HUANG Xiao-wei, LIANG Xiao-ying, QIU Ming-yue, CHEN Ying. Integrated Transcriptome and Metabolome Analysis to Explore the Carotenoid Synthesis and Metabolism Mechanism in Anoectochilus roxburghii under Different Shading Conditions [J]. Biotechnology Bulletin, 2025, 41(5): 231-243. |
| [9] | PENG Shao-zhi, WANG Deng-ke, ZHANG Xiang, DAI Xiong-ze, XU Hao, ZOU Xue-xiao. Cloning, Expression Characteristics and Functional Verification of the Pepper CaFD1 Gene [J]. Biotechnology Bulletin, 2025, 41(5): 153-164. |
| [10] | NIU Ruo-yu, GAO Zhan, XIONG Xian-peng, ZHU De, LUO Hao-tian, MA Xue-yuan, HU Guan-jing. Breeding Applications and Prospects of Wild Cotton Germplasm Resources [J]. Biotechnology Bulletin, 2025, 41(4): 21-32. |
| [11] | SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit [J]. Biotechnology Bulletin, 2025, 41(3): 190-201. |
| [12] | LIU Jie, WANG Fei, TAO Ting, ZHANG Yu-jing, CHEN Hao-ting, ZHANG Rui-xing, SHI Yu, ZHANG Yi. Overexpression of SlWRKY41 Improves the Tolerance of Tomato Seedlings to Drought [J]. Biotechnology Bulletin, 2025, 41(2): 107-118. |
| [13] | ZHOU Wen-jian, CUI Xiao-nan, SHI Wei-yang. Research Progress in the Application of DNA Methyltransferases in Epigenetics [J]. Biotechnology Bulletin, 2025, 41(2): 30-39. |
| [14] | MIAO Bai-ling, WANG Yuan-yuan, HOU Wei-wei, ZHANG Yi-ru, CHEN Juan-juan, LI Liang-jie, ZHANG He, ZHU Qing-song, DONG Xiang-xiang. Hetero-overexpression of Fragaria vescaFveBBX32 Negatively Regulates the Chlorophyll Content and Flowering Time in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2025, 41(11): 293-300. |
| [15] | ZHANG Yu-xuan, ZHANG Shi-yi, CHEN Hui-fang, CAI Kun-xiu, LI Chen-ye, YANG Jun-jie, ZHENG Tao, QIU Ming-yue, YANG You-si-yuan, CHEN Ying. Differential Accumulation of Carotenoids in Ludisia discolor under Different Light Qualities Based on Multiomics [J]. Biotechnology Bulletin, 2025, 41(10): 98-109. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||