








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 201-213.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0507
Previous Articles Next Articles
YANG Chen-xin( ), LI Meng-xiu, JIANG Tang, ZHANG Wen-e(
), LI Meng-xiu, JIANG Tang, ZHANG Wen-e( ), PAN Xue-jun
), PAN Xue-jun
Received:2025-05-17
Online:2025-12-26
Published:2026-01-06
Contact:
ZHANG Wen-e
E-mail:1073491690@qq.com;agr.wezhang@gzu.edu.cn
YANG Chen-xin, LI Meng-xiu, JIANG Tang, ZHANG Wen-e, PAN Xue-jun. Identification and Expression Analysis of Anthocyanin-associated R2R3-MYB Genes in Canna indica[J]. Biotechnology Bulletin, 2025, 41(12): 201-213.
基因名称 Gene name | 上游引物 Forward primer (5′-3′) | 下游引物 Reverse primer (5′-3′) |
|---|---|---|
| 18S | ATTCTATGGGTGGTGGTGC | CCATCCAATCGGTAGGAGC |
| CiMYB20 | AGCAGGGCTTTAACAAGGGG | GCTCCTCGGGCATCGATTTA |
| CiMYB39 | GAATGCAGGACTGCTGAGGT | GGTTGACGATGGTGTCGTCT |
| CiMYB65 | TGTTCCACCACCGCCATATC | CCAATCGGAGAACTCGCTGT |
| CiMYB116 | CGTCGTCAGTAGTACCGCTC | CTCCTCGTTGGGATCGACAG |
| CiMYB16 | GGCAGCCAACAAAAGGCTAC | GTCTCTGGATCTGCGTTGCT |
| CiMYB179 | GCTCATAGGCAACAGGTGGT | GAGCTGAGCTTTGACCGAGT |
| CiMYB8 | CCAGCAGCTCAAATCCTCCA | ACCAAGTCCTGCATGCTCTC |
| CiMYB80 | ACGGAGACTTGTTCGGTGTC | TGGCCATGTTGGGTTGTGAT |
| CiMYB21 | TTTGCTGACTTCAATGCGGC | CCTCCTCGATGAATCTCGCC |
| CiMYB171 | CGGAGGAAACTTACGGCACT | CTCCTCGGAGGCAATGAGTC |
| CiMYB68 | CCAGCCCTTGCTGTTACTGA | CCAGCAGCTCAAATCCTCCA |
Table 1 Primers for CiMYBs genes
基因名称 Gene name | 上游引物 Forward primer (5′-3′) | 下游引物 Reverse primer (5′-3′) |
|---|---|---|
| 18S | ATTCTATGGGTGGTGGTGC | CCATCCAATCGGTAGGAGC |
| CiMYB20 | AGCAGGGCTTTAACAAGGGG | GCTCCTCGGGCATCGATTTA |
| CiMYB39 | GAATGCAGGACTGCTGAGGT | GGTTGACGATGGTGTCGTCT |
| CiMYB65 | TGTTCCACCACCGCCATATC | CCAATCGGAGAACTCGCTGT |
| CiMYB116 | CGTCGTCAGTAGTACCGCTC | CTCCTCGTTGGGATCGACAG |
| CiMYB16 | GGCAGCCAACAAAAGGCTAC | GTCTCTGGATCTGCGTTGCT |
| CiMYB179 | GCTCATAGGCAACAGGTGGT | GAGCTGAGCTTTGACCGAGT |
| CiMYB8 | CCAGCAGCTCAAATCCTCCA | ACCAAGTCCTGCATGCTCTC |
| CiMYB80 | ACGGAGACTTGTTCGGTGTC | TGGCCATGTTGGGTTGTGAT |
| CiMYB21 | TTTGCTGACTTCAATGCGGC | CCTCCTCGATGAATCTCGCC |
| CiMYB171 | CGGAGGAAACTTACGGCACT | CTCCTCGGAGGCAATGAGTC |
| CiMYB68 | CCAGCCCTTGCTGTTACTGA | CCAGCAGCTCAAATCCTCCA |
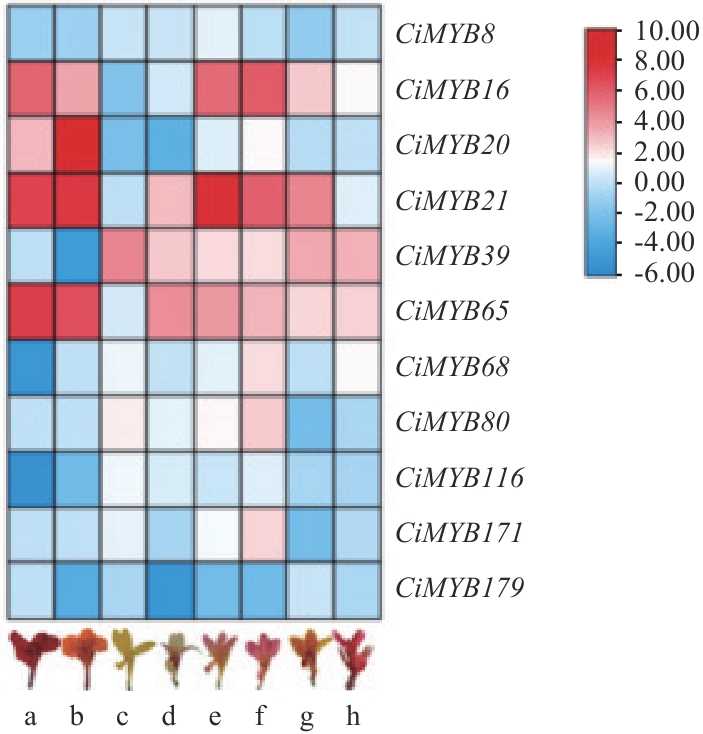
Fig. 8 Expression patterns of R2R3-MYB genes in Canna lilya: Anwang. b: Yinli. c: Progeny 1 of aquatic Canna. d: Progeny 2 of aquatic Canna. e: Progeny 3 of aquatic Canna. f: Progeny 4 of aquatic Canna. g: Progeny 1 of Canna generalis. h: Progeny 2 of Canna generalis

Fig. 9 Interaction analysis between anthocyanin-related Canna lilyThe connecting lines indicate co-expression relationships between genes, with color coding reflecting the nature of interactions. Red lines indicate significantly positive correlations (synergistic regulation), blue lines denote significantly negative correlations (repressive or antagonistic effects), and the thickness of the lines is positively correlated with the strength of co-expression or statistical confidence
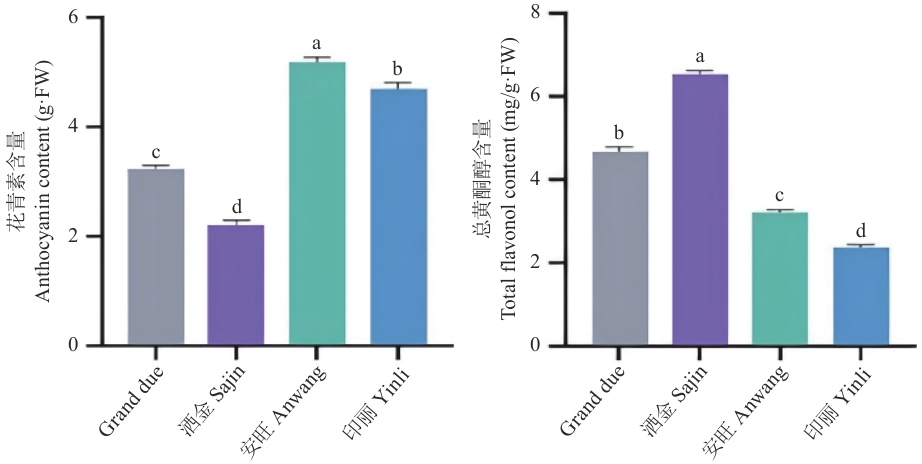
Fig. 10 Contents of total anthocyanin and total flavonol in Canna lilyDifferent lowercase letters indicate significant differences among different treatments (P<0.05). The same below
| [32] | Liu SA, Zhang HY, Meng ZL, et al. The LncNAT11-MYB11-F3'H/FLS module mediates flavonol biosynthesis to regulate salt stress tolerance in Ginkgo biloba [J]. J Exp Bot, 2025, 76(4): 1179-1201. |
| [33] | Li WF, Gao J, Ma ZH, et al. Molecular evolution and expression assessment of DFRs in apple [J]. Chem Biol Technol Agric, 2023, 10(1): 98. |
| [34] | Zhang L, Ma JN. Promoter cloning of VcCHS gene from blueberries and selection of its transcription factors [J]. Hortic Environ Biotechnol, 2025. DOI: https://doi.org/10.1007/s13580-025-00694-y . |
| [35] | Chen DZ, Wang CC, Liu Y, et al. Systematic identification of R2R3-MYB S6 subfamily genes in Brassicaceae and its role in anthocyanin biosynthesis in Brassica crops [J]. BMC Plant Biol, 2025, 25(1): 290. |
| [36] | Bhatia C, Gaddam SR, Pandey A, et al. COP1 mediates light-dependent regulation of flavonol biosynthesis through HY5 in Arabidopsis [J]. Plant Sci, 2021, 303: 110760. |
| [37] | Shi LY, Chen X, Wang K, et al. MrMYB6 from Chinese bayberry (Myrica rubra) negatively regulates anthocyanin and proanthocyanidin accumulation [J]. Front Plant Sci, 2021, 12: 685654. |
| [38] | Duan AQ, Tan SS, Deng YJ, et al. Genome-wide identification and evolution analysis of R2R3-MYB gene family reveals S6 subfamily R2R3-MYB transcription factors involved in anthocyanin biosynthesis in carrot [J]. Int J Mol Sci, 2022, 23(19): 11859. |
| [39] | Roy S, Tripathi AM, Yadav A, et al. Identification and expression analyses of miRNAs from two contrasting flower color cultivars of Canna by deep sequencing [J]. PLoS One, 2016, 11(1): e0147499. |
| [1] | Qi YT, Gu CH, Wang XJ, et al. Identification of the Eutrema salsugineum EsMYB90 gene important for anthocyanin biosynthesis [J]. BMC Plant Biol, 2020, 20(1): 186. |
| [2] | Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis [J]. Trends Plant Sci, 2010, 15(10): 573-581. |
| [3] | Nakatsuka T, Haruta KS, Pitaksutheepong C, et al. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers [J]. Plant Cell Physiol, 2008, 49(12): 1818-1829. |
| [4] | Yan HL, Pei XN, Zhang H, et al. MYB-mediated regulation of anthocyanin biosynthesis [J]. Int J Mol Sci, 2021, 22(6): 3103. |
| [5] | Poudel PR, Azuma A, Kobayashi S, et al. VvMYBAs induce expression of a series of anthocyanin biosynthetic pathway genes in red grapes (Vitis vinifera L.) [J]. Sci Hortic, 2021, 283: 110121. |
| [6] | Laitinen RAE, Ainasoja M, Broholm SK, et al. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida [J]. J Exp Bot, 2008, 59(13): 3691-3703. |
| [7] | Yamagishi M, Shimoyamada Y, Nakatsuka T, et al. Two R2R3-MYB genes, homologs of Petunia AN2 regulate anthocyanin biosyntheses in flower Tepals, tepal spots and leaves of Asiatic hybrid lily [J]. Plant Cell Physiol, 2010, 51(3): 463-474. |
| [8] | Zheng YL, Chen YD, Liu ZG, et al. Important roles of key genes and transcription factors in flower color differences of Nicotiana alata [J]. Genes, 2021, 12(12): 1976. |
| [9] | 黄国涛, 欧阳底梅, 向其柏, 等. 美人蕉属品种分类研究 [J]. 南京林业大学学报: 自然科学版, 2005, 29(4): 20-24. |
| Huang GT, Ouyang DM, Xiang QB, et al. Studies on classification for cultivars of Canna L [J]. J Nanjing For Univ, 2005, 29(4): 20-24. | |
| [10] | Tripathi AM, Niranjan A, Roy S. Global gene expression and pigment analysis of two contrasting flower color cultivars of Canna [J]. Plant Physiol Biochem, 2018, 127: 1-10. |
| [11] | Liu MM, Li C, Jiang T, et al. Chromosome-scale genome assembly provides insights into flower coloration mechanisms of Canna indica [J]. Int J Biol Macromol, 2023, 251: 126148. |
| [12] | 肖伟军, 任小玉, 杨芯蕊, 等. 香蕉R 2R3-MYB基因家族鉴定及在果实发育中的表达分析 [J/OL]. 分子植物育种, 2024. . |
| Xiao WJ, Ren XY, Yang XR, et al. Genomic identification of the R 2R3-MYB gene family in banana and expression analysis during fruit development [J/OL]. Mol Plant Breed, 2024. . | |
| [13] | Song HY, Duan ZH, Wang Z, et al. Genome-wide identification, expression pattern and subcellular localization analysis of the JAZ gene family in Toona ciliata [J]. Ind Crops Prod, 2022, 178: 114582. |
| [14] | Chen GQ, He WZ, Guo XX, et al. Genome-wide identification, classification and expression analysis of the MYB transcription factor family in Petunia [J]. Int J Mol Sci, 2021, 22(9): 4838. |
| [15] | Tian SN, Ali MM, Ke ML, et al. Novel R2R3-MYB transcription factor LhMYB1 promotes anthocyanin accumulation in Lilium concolor var. pulchellum [J]. Horticulturae, 2024, 10(5): 509. |
| [16] | Stracke R, Ishihara H, Huep G, et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling [J]. Plant J, 2007, 50(4): 660-677. |
| [17] | Zhu ZP, Yu JX, Liu FF, et al. AeWRKY32 from okra regulates anthocyanin accumulation and cold tolerance in Arabidopsis [J]. J Plant Physiol, 2023, 287: 154062. |
| [18] | Chen LH, Hu B, Qin YH, et al. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors [J]. Plant Physiol Biochem, 2019, 136: 178-187. |
| [19] | 赵婷. 美人蕉属植物耐涝性评价及对淹水胁迫的形态生理与基因表达差异响应 [D]. 贵阳: 贵州大学, 2022. |
| Zhao T. Evaluation of waterlogging tolerance of Canna plants and its response to morphophysiological, physiological and gene expression differences under waterlogging stress [D]. Guiyang: Guizhou University, 2022. | |
| [20] | Shi YF, Lu TR, Lai SY, et al. Rosa rugosa R2R3-MYB transcription factors RrMYB12 and RrMYB111 regulate the accumulation of flavonols and anthocyanins [J]. Front Plant Sci, 2024, 15: 1477278. |
| [21] | Zhu J, Wang YZ, Zhou X, et al. Characterization of three novel R2R3-MYB transcription factors PrMYBi (1-3) repressing the anthocyanin biosynthesis in tree peony [J]. Hortic Plant J, 2024. DOI: https://doi.org/10.1016/j.hpj.2024.08.005 . |
| [22] | 刘雅芝, 邓惠敏, 彭嘉慧, 等. 铁十字秋海棠R2R3-MYB基因家族的鉴定及表达分析 [J]. 南方农业学报, 2024, 55(9): 2665-2678. |
| Liu YZ, Deng HM, Peng JH, et al. Identification and expression analysis of R2R3-MYB gene family in Begonia masoniana [J]. J South Agric, 2024, 55(9): 2665-2678. | |
| [23] | Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana [J]. Curr Opin Plant Biol, 2001, 4(5): 447-456. |
| [24] | Hu XM, Liang ZH, Sun TX, et al. The R2R3-MYB transcriptional repressor TgMYB4 negatively regulates anthocyanin biosynthesis in tulips (Tulipa gesneriana L.) [J]. Int J Mol Sci, 2024, 25(1): 563. |
| [25] | Alabd A, Ahmad M, Zhang X, et al. Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear [J]. Hortic Res, 2022, 9: uhac199. |
| [26] | Hsu CC, Chen YY, Tsai WC, et al. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp [J]. Plant Physiol, 2015, 168(1): 175-191. |
| [27] | Gu ZY, Zhu J, Hao Q, et al. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa) [J]. Plant Cell Physiol, 2019, 60(3): 599-611. |
| [28] | Wang CP, Li Y, Wang N, et al. An efficient CRISPR/Cas9 platform for targeted genome editing in rose (Rosa hybrida) [J]. J Integr Plant Biol, 2023, 65(4): 895-899. |
| [29] | Yang L, Wang G, Liu TT, et al. The MYB transcription factor RtAN2 promotes anthocyanin accumulation over proanthocyanidin during the fruit ripening of rose myrtle berries [J]. Ind Crops Prod, 2025, 224: 120433. |
| [30] | Cui C, Zhang K, Chai L, et al. Unraveling the mechanism of flower color variation in Brassica napus by integrated metabolome and transcriptome analyses [J]. Front Plant Sci, 2024, 15: 1419508. |
| [31] | Zuluaga DL, Gonzali S, Loreti E, et al. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants [J]. Funct Plant Biol, 2008, 35(7): 606-618. |
| [1] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [2] | LI Yu-zhen, LI Meng-dan, ZHANG Wei, PENG Ting. Functional Study of RmEXPB2 Genein Rosa multiflora Based on the Identification of the Expansin Gene Family in Rosa sp. [J]. Biotechnology Bulletin, 2025, 41(9): 182-194. |
| [3] | ZHANG Ze, YANG Xiu-li, NING Dong-xian. Identification of 4CL Gene Family in Arachis hypogaea L. and Expression Analysis in Response to Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(7): 117-127. |
| [4] | GUO Tao, AI Li-jiao, ZOU Shi-hui, ZHOU Ling, LI Xue-mei. Functional Study of CjRAV1 from Camellia japonica in Regulating Flowering Delay [J]. Biotechnology Bulletin, 2025, 41(6): 208-217. |
| [5] | ZHOU Yi, LIU Yong-bo. Research Progress in the Mechanisms and Functions of Gene Loss in Genome Evolution [J]. Biotechnology Bulletin, 2025, 41(6): 38-48. |
| [6] | LIU Hong-li, MA Yi-dan, WANG Wan-ru, YANG Ya-ru, HE Dan, LIU Yi-ping, KONG De-zheng. Cloning and Functional Analysis of NnCYP707A1 Gene from Lotus [J]. Biotechnology Bulletin, 2025, 41(5): 197-207. |
| [7] | SONG Hui-yang, SU Bao-jie, LI Jing-hao, MEI Chao, SONG Qian-na, CUI Fu-zhu, FENG Rui-yun. Cloning and Functional Analysis of the StAS2-15 Gene in Potato under Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 119-128. |
| [8] | LIU Yuan, ZHAO Ran, LU Zhen-fang, LI Rui-li. Research Progress in the Biological Metabolic Pathway and Functions of Plant Carotenoids [J]. Biotechnology Bulletin, 2025, 41(5): 23-31. |
| [9] | TIAN Qin, LIU Kui, WU Xiang-wei, JI Yuan-yuan, CAO Yi-bo, ZHANG Ling-yun. Functional Study of Transcription Factor VcMYB17 in Regulating Drought Tolerance in Blueberry [J]. Biotechnology Bulletin, 2025, 41(4): 198-210. |
| [10] | LI Bin, SU Xiang-ping, LIU Chang, WANG Yu-bing, ZHANG Yong-hong, ZHOU Chao, XU Qing. Chloroplast Genome Characteristics and Phylogenetic Analysis of Scrophulariaceae [J]. Biotechnology Bulletin, 2025, 41(3): 240-254. |
| [11] | MIAO Hao-xiang, ZHANG Ying, GUO Shi-peng, ZHANG Jian. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yielding γ-aminobutyric Acid-producing Lactobacillus brevis TCCC13007 [J]. Biotechnology Bulletin, 2025, 41(11): 166-176. |
| [12] | LIU Zi-qi, ZHONG Pei, LI Qin, GUO Cheng, ZHANG Yan-mei, ZHANG Nai-feng, TU Yan, DIAO Qi-yu, BI Yan-liang. Application and Progress of CRISPR/Cas9 Technology in Probiotic Editing [J]. Biotechnology Bulletin, 2025, 41(11): 89-99. |
| [13] | QIN Wen-jun, XIONG Yan-jie, ZHAO Ran, MA Xiao-ran, YE Xiao-meng, SONG Jiang-hua. Identification and Expression Analysis of the ARF Gene Family in Cabbage under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(10): 253-263. |
| [14] | DU Pin-ting, WU Guo-jiang, WANG Zhen-guo, LI Yan, ZHOU Wei, ZHOU Ya-xing. Identification and Expression Analysis of CPP Gene Family in Sorghum [J]. Biotechnology Bulletin, 2025, 41(1): 132-142. |
| [15] | LI Yong-hui, BAO Xing-xing, DUAN Yi-ke, ZHAO Yun-xia, YU Xiang-li, CHEN Yao, ZHANG Yan-zhao. Genome-wide Identification and Expression Features Analysis of the bZIP Family in Rhododendron henanense subsp. lingbaoense [J]. Biotechnology Bulletin, 2024, 40(8): 186-198. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||