








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 47-61.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0713
Previous Articles Next Articles
ZHOU Si-yan1,2,3( ), DING Wei-quan2, DONG Ping2, WENG Han-zhi1,2,3, XU Rui-rui1,2,3, KANG Zhen1,2,3(
), DING Wei-quan2, DONG Ping2, WENG Han-zhi1,2,3, XU Rui-rui1,2,3, KANG Zhen1,2,3( )
)
Received:2025-07-02
Online:2025-11-26
Published:2025-12-09
Contact:
KANG Zhen
E-mail:b7240201092@stu.jiangnan.edu.cn;zkang@jiangnan.edu.cn
ZHOU Si-yan, DING Wei-quan, DONG Ping, WENG Han-zhi, XU Rui-rui, KANG Zhen. Advances in Synthetic Biology Platform Development and Application for Escherichiacoli Nissle 1917[J]. Biotechnology Bulletin, 2025, 41(11): 47-61.
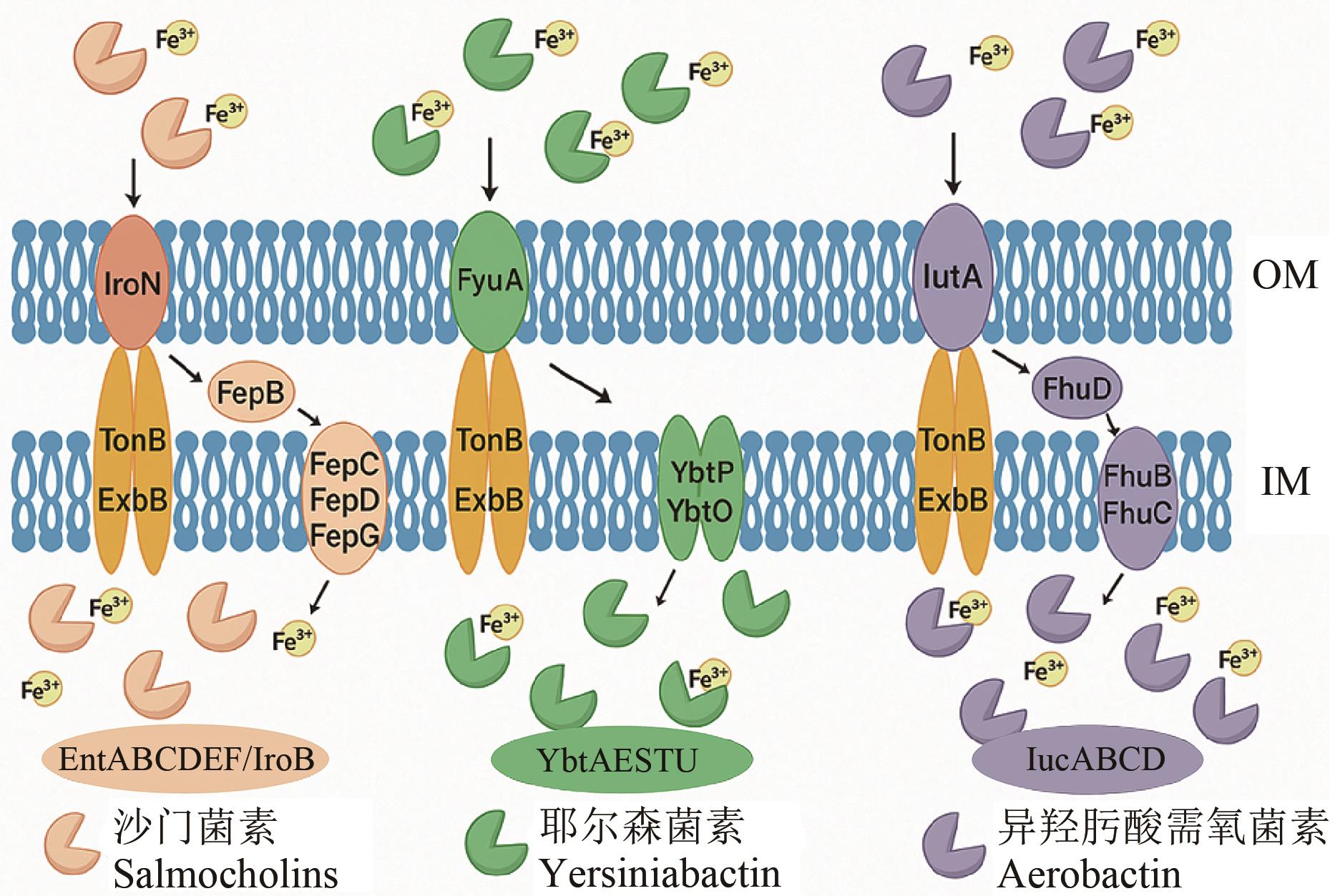
Fig. 3 Mechanism of iron uptake by EcN through related proteins and systems on the outer membrane (OM) and inner membrane (IM)As shown in the figure, EcN employs multiple pathways to take up iron bound to different substances, utilizing various proteins such as EntABCDEF and IroB. Proteins like TonB and ExbB play a role in transmembrane transport
工具 Tool | 类型 Type | 用途 Aplication | 文献来源 Reference |
|---|---|---|---|
| 启动子 | 组成型 | 以J23100启动子表达gabB基因,提高GABA的产量 | [ |
| 通过PfnrS启动子表达phaA、phaB、tesB、mailKE基因,提高3HB的产量 | [ | ||
| 化学诱导 | T7表达系统表达铁氧还蛋白氧化还原酶PcyA | [ | |
| 通过T7表达系统表达基因产生血红素代谢物 | [ | ||
| 通过T7表达系统同时合成磺基转移酶和肝素前体 | [ | ||
| 通过lac启动子表达kfiC基因控制荚膜 | [ | ||
| 光诱导 | 通过蓝光负责启动子表达Ag43、IL-10、TNFα、FlaB,进行精准治疗 | [ | |
| 红光遗传电路裂解细胞释放组成表达的Ex-4-aa | [ | ||
| CRISPR/Cas | CRISPRi | 抑制ccdAB和hipAB基因表达平衡生物膜的定植和毒性 | [ |
| csgD基因的抑制作用 | [ | ||
| 基因编辑 | 敲除frdA、ldhA、adhE和pta基因,提高GABA的产量 | [ | |
| 敲除内源隐性质粒 | [ | ||
| 作为一种靶向抗生素耐药基因的抗菌药物 | [ | ||
| “自杀”系统 | 光诱导 | 触发毒素蛋白CcdB的积累和抗毒素蛋白CcdA的减少 | [ |
| CRISPR | 引导Cas9到达切割基因组的目标序列 | [ | |
| 群体感应 | 触发lysin的表达,释放蛋白S5和DspB | [ | |
| lux启动子驱动的luxI转录,以及噬菌体衍生的裂解基因φX174E | [ |
Table 1 Genetic tools for EcN
工具 Tool | 类型 Type | 用途 Aplication | 文献来源 Reference |
|---|---|---|---|
| 启动子 | 组成型 | 以J23100启动子表达gabB基因,提高GABA的产量 | [ |
| 通过PfnrS启动子表达phaA、phaB、tesB、mailKE基因,提高3HB的产量 | [ | ||
| 化学诱导 | T7表达系统表达铁氧还蛋白氧化还原酶PcyA | [ | |
| 通过T7表达系统表达基因产生血红素代谢物 | [ | ||
| 通过T7表达系统同时合成磺基转移酶和肝素前体 | [ | ||
| 通过lac启动子表达kfiC基因控制荚膜 | [ | ||
| 光诱导 | 通过蓝光负责启动子表达Ag43、IL-10、TNFα、FlaB,进行精准治疗 | [ | |
| 红光遗传电路裂解细胞释放组成表达的Ex-4-aa | [ | ||
| CRISPR/Cas | CRISPRi | 抑制ccdAB和hipAB基因表达平衡生物膜的定植和毒性 | [ |
| csgD基因的抑制作用 | [ | ||
| 基因编辑 | 敲除frdA、ldhA、adhE和pta基因,提高GABA的产量 | [ | |
| 敲除内源隐性质粒 | [ | ||
| 作为一种靶向抗生素耐药基因的抗菌药物 | [ | ||
| “自杀”系统 | 光诱导 | 触发毒素蛋白CcdB的积累和抗毒素蛋白CcdA的减少 | [ |
| CRISPR | 引导Cas9到达切割基因组的目标序列 | [ | |
| 群体感应 | 触发lysin的表达,释放蛋白S5和DspB | [ | |
| lux启动子驱动的luxI转录,以及噬菌体衍生的裂解基因φX174E | [ |
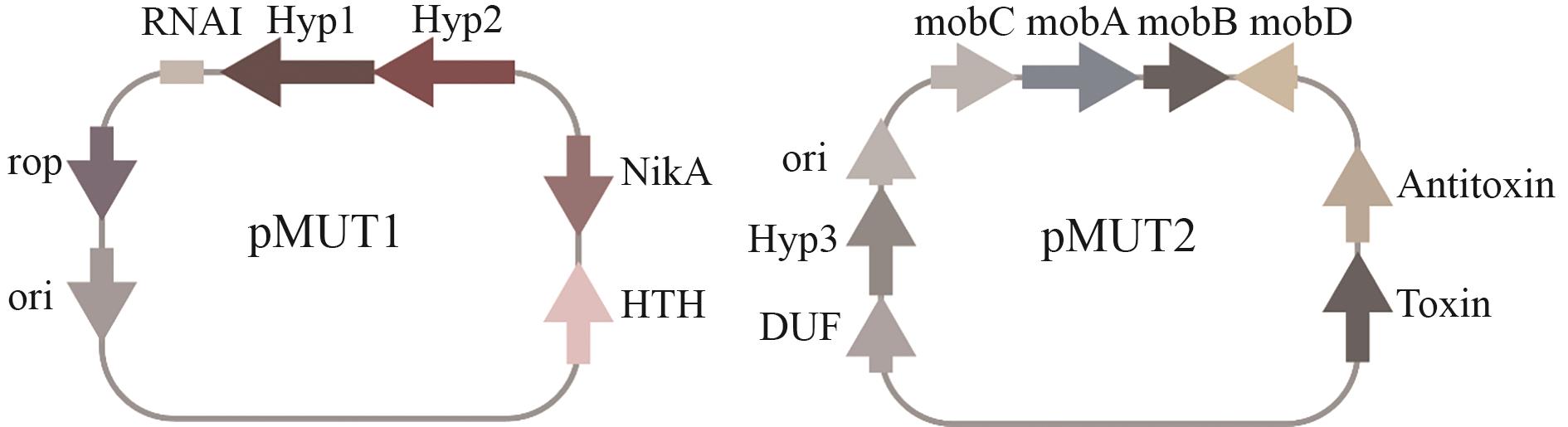
Fig. 4 Cryptic plasmids (pMUT1and pMUT2) in EcNGenetic structures and related elements of two cryptic plasmids pMUT1 and pMUT2. pMUT1 contains genes or elements such as Rop, ori, NikA, HTH, Hyp1, and Hyp2, and also involves RNAI-related components; pMUT2 contains genes or elements including ori, MobC, MobA, MobB, MobD, Antitoxin, Toxin, Hyp3, and DUF
疾病 Disease | EcN的作用 Effect of EcN | 动物模型/细胞系 Animal model/Cell line | 参考文献 References |
|---|---|---|---|
| 多重致敏 | Th2淋巴细胞减少 | 鼠 | [ |
| 感染 | 免疫调节 | 猪 | [ |
| 过敏性哮喘 | 抑制T2和ILC2的激活 | 鼠 | [ |
| 志贺氏菌感染 | 干扰噬菌体感染易感大肠杆菌 | 大肠杆菌 | [ |
| 空肠弯曲杆菌感染 | 调节保护性先天免疫相关基因的表达 | HT-29细胞 | [ |
| 非传染性葡萄膜炎 | 调节肠眼轴 | 鼠 | [ |
| 肠上皮屏障功能障碍 | 维持与TJ结构相关的亚细胞定位 | T-84和Caco-2细胞 | [ |
| 人轮状病毒感染 | 增强先天和B细胞免疫反应 | 猪 | [ |
| 结肠癌 | 通过调节信号通路诱导细胞凋亡 | HT-29细胞 | [ |
| IBD | 调节特定mRNA的表达 | 鼠 | [ |
| 呼吸道合胞病毒 | 逆转肠道生态失调 | 鼠 | [ |
| 抗生素不适 | 阻断病理肠脑回路 | 鼠 | [ |
| 慢性细菌性前列腺炎 | 左氧氟沙星联合治疗 | 人体 | [ |
| 肠道炎症性/感染性腹泻 | 外膜囊泡调节免疫细胞的功能 | RAW 264.7细胞 | [ |
| 苯丙酮尿症 | 降解丙氨酸 | 人体 | [ |
| 口腔癌 | 检测乳酸 | 人体 | [ |
Table 2 Clinical application of EcN
疾病 Disease | EcN的作用 Effect of EcN | 动物模型/细胞系 Animal model/Cell line | 参考文献 References |
|---|---|---|---|
| 多重致敏 | Th2淋巴细胞减少 | 鼠 | [ |
| 感染 | 免疫调节 | 猪 | [ |
| 过敏性哮喘 | 抑制T2和ILC2的激活 | 鼠 | [ |
| 志贺氏菌感染 | 干扰噬菌体感染易感大肠杆菌 | 大肠杆菌 | [ |
| 空肠弯曲杆菌感染 | 调节保护性先天免疫相关基因的表达 | HT-29细胞 | [ |
| 非传染性葡萄膜炎 | 调节肠眼轴 | 鼠 | [ |
| 肠上皮屏障功能障碍 | 维持与TJ结构相关的亚细胞定位 | T-84和Caco-2细胞 | [ |
| 人轮状病毒感染 | 增强先天和B细胞免疫反应 | 猪 | [ |
| 结肠癌 | 通过调节信号通路诱导细胞凋亡 | HT-29细胞 | [ |
| IBD | 调节特定mRNA的表达 | 鼠 | [ |
| 呼吸道合胞病毒 | 逆转肠道生态失调 | 鼠 | [ |
| 抗生素不适 | 阻断病理肠脑回路 | 鼠 | [ |
| 慢性细菌性前列腺炎 | 左氧氟沙星联合治疗 | 人体 | [ |
| 肠道炎症性/感染性腹泻 | 外膜囊泡调节免疫细胞的功能 | RAW 264.7细胞 | [ |
| 苯丙酮尿症 | 降解丙氨酸 | 人体 | [ |
| 口腔癌 | 检测乳酸 | 人体 | [ |
产物 Product | 策略 Strategy | 产量 Yield | 参考文献 Reference |
|---|---|---|---|
| β-丙氨酸 | 表达天冬氨酸转移酶,增加富马酸的供应,增强前体途径 | 11.9 g/L | [ |
| L-精氨酸 | 删除负调控基因,插入抗反馈酶基因 | - | [ |
| 3-羟基丁酸 | 将合酶基因整合基因组 | 0.6 g/L | [ |
| 丁酸 | 异源表达BCD和BUT基因 | 297 mg/L | [ |
| 丁酸盐 | 葡萄糖途径整合丁基辅酶A | 20 mmol/L | [ |
| 5-氨基乙酰丙酸 | 过表达合酶,抑制降解途径 | 300 mg/L | [ |
| 胰高血糖素样肽-1 | 过表达编码基因 | - | [ |
| 血红素衍生分子 | 在EcN基因组上整合T7表达系统,表达含血红素蛋白 | - | [ |
| γ-氨基丁酸 | 使用无抗质粒作为载体 | 17.9 g/L | [ |
| 细菌纤维素 | 过表达纤维素合酶 | 1.94 g/L | [ |
| Omega-3 | 过表达合酶基因簇 | 31.36 mg/L | [ |
| 肝素前体 | 强化前体途径,过表达合酶,调节转运蛋白 | 12.2 g/L | [ |
Table 3 Industrial application of EcN
产物 Product | 策略 Strategy | 产量 Yield | 参考文献 Reference |
|---|---|---|---|
| β-丙氨酸 | 表达天冬氨酸转移酶,增加富马酸的供应,增强前体途径 | 11.9 g/L | [ |
| L-精氨酸 | 删除负调控基因,插入抗反馈酶基因 | - | [ |
| 3-羟基丁酸 | 将合酶基因整合基因组 | 0.6 g/L | [ |
| 丁酸 | 异源表达BCD和BUT基因 | 297 mg/L | [ |
| 丁酸盐 | 葡萄糖途径整合丁基辅酶A | 20 mmol/L | [ |
| 5-氨基乙酰丙酸 | 过表达合酶,抑制降解途径 | 300 mg/L | [ |
| 胰高血糖素样肽-1 | 过表达编码基因 | - | [ |
| 血红素衍生分子 | 在EcN基因组上整合T7表达系统,表达含血红素蛋白 | - | [ |
| γ-氨基丁酸 | 使用无抗质粒作为载体 | 17.9 g/L | [ |
| 细菌纤维素 | 过表达纤维素合酶 | 1.94 g/L | [ |
| Omega-3 | 过表达合酶基因簇 | 31.36 mg/L | [ |
| 肝素前体 | 强化前体途径,过表达合酶,调节转运蛋白 | 12.2 g/L | [ |
| [1] | Sonnenborn U, Schulze J. The non-pathogenic Escherichia coli strain Nissle 1917-features of a versatile probiotic [J]. Microb Ecol Health Dis, 2009, 21(3/4): 122-158. |
| [2] | Alvarez CS, Giménez R, Cañas MA, et al. Extracellular vesicles and soluble factors secreted by Escherichia coli Nissle 1917 and ECOR63 protect against enteropathogenic E. coli-induced intestinal epithelial barrier dysfunction [J]. BMC Microbiol, 2019, 19(1): 166. |
| [3] | Alizadeh S, Esmaeili A, Omidi Y. Anti-cancer properties of Escherichia coli Nissle 1917 against HT-29 colon cancer cells through regulation of Bax/Bcl-xL and AKT/PTEN signaling pathways [J]. Iran J Basic Med Sci, 2020, 23(7): 886-893. |
| [4] | Behrouzi A, Mazaheri H, Falsafi S, et al. Intestinal effect of the probiotic Escherichia coli strain Nissle 1917 and its OMV [J]. J Diabetes Metab Disord, 2020, 19(1): 597-604. |
| [5] | Praveschotinunt P, Duraj-Thatte AM, Gelfat I, et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut [J]. Nat Commun, 2019, 10: 5580. |
| [6] | Rodríguez-Nogales A, Algieri F, Garrido-Mesa J, et al. The administration of Escherichia coli nissle 1917 ameliorates development of DSS-induced colitis in mice [J]. Front Pharmacol, 2018, 9: 468. |
| [7] | Ou BM, Yang Y, Tham WL, et al. Genetic engineering of probiotic Escherichia coli Nissle 1917 for clinical application [J]. Appl Microbiol Biotechnol, 2016, 100(20): 8693-8699. |
| [8] | Sonnenborn U. Escherichia coli strain Nissle 1917—from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties [J]. FEMS Microbiol Lett, 2016, 363(19): fnw212. |
| [9] | Irrgang K, Sonnenborn U. The Historical Development of Mutaflor Therapy [M]. Ardeypharm, 1988. |
| [10] | Patzer SI, Baquero MR, Bravo D, et al. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN [J]. Microbiology, 2003, 149(9): 2557-2570. |
| [11] | Deriu E, Liu JZ, Pezeshki M, et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron [J]. Cell Host Microbe, 2013, 14(1): 26-37. |
| [12] | Ukena SN, Singh A, Dringenberg U, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity [J]. PLoS One, 2007, 2(12): e1308. |
| [13] | Altenhoefer A, Oswald S, Sonnenborn U, et al. The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens [J]. FEMS Immunol Med Microbiol, 2004, 40(3): 223-229. |
| [14] | Wehkamp J, Harder J, Wehkamp K, et al. NF-κB- and AP-1-Mediated induction of human beta Defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium [J]. Infect Immun, 2004, 72(10): 5750-5758. |
| [15] | Reister M, Hoffmeier K, Krezdorn N, et al. Complete genome sequence of the Gram-negative probiotic Escherichia coli strain Nissle 1917 [J]. J Biotechnol, 2014, 187: 106-107. |
| [16] | Henker J, Laass M, Blokhin BM, et al. The probiotic Escherichia coli strain Nissle 1917 (EcN) stops acute diarrhoea in infants and toddlers [J]. Eur J Pediatr, 2007, 166(4): 311-318. |
| [17] | Scaldaferri F, Gerardi V, Mangiola F, et al. Role and mechanisms of action of Escherichia coli Nissle 1917 in the maintenance of remission in ulcerative colitis patients: an update [J]. World J Gastroenterol, 2016, 22(24): 5505. |
| [18] | Behnsen J, Deriu E, Sassone-Corsi M, et al. Probiotics: properties, examples, and specific applications [J]. Cold Spring Harb Perspect Med, 2013, 3(3): a010074. |
| [19] | Gasbarrini G, Bonvicini F, Gramenzi A. Probiotics history [J]. J Clin Gastroenterol, 2016, 50(): S116-S119. |
| [20] | Hu MM, Wu XL, Luo M, et al. Lactobacillus rhamnosus FLRH93 protects against intestinal damage in mice induced by 5-fluorouracil [J]. J Dairy Sci, 2020, 103(6): 5003-5018. |
| [21] | Grozdanov L, Raasch C, Schulze J, et al. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917 [J]. J Bacteriol, 2004, 186(16): 5432-5441. |
| [22] | Sassone-Corsi M, Nuccio SP, Liu H, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut [J]. Nature, 2016, 540(7632): 280-283. |
| [23] | Mortzfeld BM, Palmer JD, et al. Microcin MccI47 selectively inhibits enteric bacteria and reduces carbapenem-resistant Klebsiella pneumoniae colonization in vivo when administered viaan engineered live biotherapeutic [J]. Gut Microbes, 2022, 14: 2127633. |
| [24] | Barbaro MR, Fuschi D, Cremon C, et al. Escherichia coli Nissle 1917 restores epithelial permeability alterations induced by irritable bowel syndrome mediators [J]. Neurogastroenterol Motil, 2018, 30(8): e13388. |
| [25] | Henker J, Laass MW, Blokhin BM, et al. Probiotic Escherichia coli nissle 1917 versus placebo for treating diarrhea of greater than 4 days duration in infants and toddlers [J]. Pediatr Infect Dis J, 2008, 27(6): 494-499. |
| [26] | Zhao ZJ, Xu SM, Zhang WY, et al. Probiotic Escherichia coli NISSLE 1917 for inflammatory bowel disease applications [J]. Food Funct, 2022, 13(11): 5914-5924. |
| [27] | Yang XY, Yang JL, Ye ZH, et al. Physiologically inspired mucin coated Escherichia coli Nissle 1917 enhances biotherapy by regulating the pathological microenvironment to improve intestinal colonization [J]. ACS Nano, 2022, 16(3): 4041-4058. |
| [28] | Geervliet M, de Vries H, Jansen CA, et al. Effects of Escherichia coli Nissle 1917 on the porcine gut microbiota, intestinal epithelium and immune system in early life [J]. Front Microbiol, 2022, 13: 842437. |
| [29] | Haneishi Y, Furuya Y, Hasegawa M, et al. Inflammatory bowel diseases and gut microbiota [J]. Int J Mol Sci, 2023, 24(4): 3817. |
| [30] | Jia K, Tong X, Wang R, et al. The clinical effects of probiotics for inflammatory bowel disease: a meta-analysis [J]. Medicine, 2018, 97(51): e13792. |
| [31] | Park J, Kim DH, Kim S, et al. Anti-inflammatory properties of Escherichia coli Nissle 1917 in a murine colitis model [J]. Intest Res, 2021, 19(4): 478-481. |
| [32] | Nougayrède JP, Chagneau CV, Motta JP, et al. A toxic friend: genotoxic and mutagenic activity of the probiotic strain Escherichia coli Nissle 1917 [J]. mSphere, 2021, 6(4): e00624-21. |
| [33] | Wang Y, Fu K. Genotoxins: the mechanistic links between Escherichia coli and colorectal cancer [J]. Cancers, 2023, 15(4): 1152. |
| [34] | Janosch D, Dubbert S, Eiteljörge K, et al. Anti-genotoxic and anti-mutagenic activity of Escherichia coli Nissle 1917 as assessed by in vitro tests [J]. Benef Microbes, 2019, 10(4): 449-462. |
| [35] | Dubbert S, Klinkert B, Schimiczek M, et al. No genotoxicity is detectable for Escherichia coli strain Nissle 1917 by standard in vitro and in vivo tests [J]. EuJMI, 2020, 10(1): 11-19. |
| [36] | Tóth AG, Csabai I, Judge MF, et al. Mobile antimicrobial resistance genes in probiotics [J]. Antibiotics, 2021, 10(11): 1287. |
| [37] | Iftekhar A, Berger H, Bouznad N, et al. Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells [J]. Nat Commun, 2021, 12: 1003. |
| [38] | Merenstein D, Pot B, Leyer G, et al. Emerging issues in probiotic safety: 2023 perspectives [J]. Gut Microbes, 2023, 15: 2185034. |
| [39] | Zhao LL, Yin GB, Zhang YL, et al. A comparative study on the genomes, transcriptomes, and metabolic properties of Escherichia coli strains Nissle 1917, BL21(DE3), and MG1655 [J]. Eng Microbiol, 2022, 2(1): 100012. |
| [40] | Xue CF, Ting WW, Juo JJ, et al. New insight into acid-resistant enzymes from natural mutations of Escherichia coli Nissle 1917 [J]. Enzyme Microb Technol, 2024, 181: 110526. |
| [41] | Lan YJ, Tan SI, Cheng SY, et al. Development of Escherichia coli Nissle 1917 derivative by CRISPR/Cas9 and application for gamma-aminobutyric acid (GABA) production in antibiotic-free system [J]. Biochem Eng J, 2021, 168: 107952. |
| [42] | Yan X, Liu XY, Zhang D, et al. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis [J]. Cell Mol Immunol, 2021, 18(10): 2344-2357. |
| [43] | Zhou SY, Zhao LL, Zuo WJ, et al. Minimizing endogenous cryptic plasmids to construct antibiotic-free expression systems for Escherichia coli Nissle 1917 [J]. Synth Syst Biotechnol, 2024, 9(1): 165-175. |
| [44] | Fiege K, Frankenberg-Dinkel N. Construction of a new T7 promoter compatible Escherichia coli Nissle 1917 strain for recombinant production of heme-dependent proteins [J]. Microb Cell Fact, 2020, 19(1): 190. |
| [45] | Robinson EA, Frankenberg-Dinkel N, Xue FT, et al. Recombinant production of biliverdin IXβ and δ isomers in the T7 promoter compatible Escherichia coli nissle [J]. Front Microbiol, 2021, 12: 787609. |
| [46] | Li XM, Yu YY, Tang JQ, et al. The construction of a dual-functional strain that produces both polysaccharides and sulfotransferases [J]. Biotechnol Lett, 2021, 43(9): 1831-1844. |
| [47] | Effendi SSW, Ng IS. Reprogramming T7RNA polymerase in Escherichia coli Nissle 1917 under specific lac operon for efficient p-coumaric acid production [J]. ACS Synth Biol, 2022, 11(10): 3471-3481. |
| [48] | Harimoto T, Hahn J, Chen YY, et al. A programmable encapsulation system improves delivery of therapeutic bacteria in mice [J]. Nat Biotechnol, 2022, 40(8): 1259-1269. |
| [49] | Cui MH, Pang GJ, Zhang T, et al. Optotheranostic nanosystem with phone visual diagnosis and optogenetic microbial therapy for ulcerative colitis at-home care [J]. ACS Nano, 2021, 15(4): 7040-7052. |
| [50] | Cui MH, Sun T, Li SB, et al. NIR light-responsive bacteria with live bio-glue coatings for precise colonization in the gut [J]. Cell Rep, 2021, 36(11): 109690. |
| [51] | Jayaraman P, Devarajan K, Chua TK, et al. Blue light-mediated transcriptional activation and repression of gene expression in bacteria [J]. Nucleic Acids Res, 2016, 44(14): 6994-7005. |
| [52] | Pan HZ, Li LY, Pang GJ, et al. Engineered NIR light-responsive bacteria as anti-tumor agent for targeted and precise cancer therapy [J]. Chem Eng J, 2021, 426: 130842. |
| [53] | Zhu XQ, Chen SH, Hu XW, et al. Near-infrared nano-optogenetic activation of cancer immunotherapy via engineered bacteria [J]. Adv Mater, 2023, 35(8): 2207198. |
| [54] | Fernandez-Rodriguez J, Moser F, Song M, et al. Engineering RGB color vision into Escherichia coli [J]. Nat Chem Biol, 2017, 13(7): 706-708. |
| [55] | Mulvaney CA, Duarte GS, Menon S, et al. GLP-1 receptor agonists for Parkinson’s disease [J]. Cochrane Database Syst Rev, 2020, (7): CD012990. |
| [56] | Zhang XY, Pang GJ, Sun T, et al. A red light-controlled probiotic bio-system for in situ gut-brain axis regulation [J]. Biomaterials, 2023, 294: 122005. |
| [57] | Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR-Cas systems [J]. Nat Rev Microbiol, 2015, 13(11): 722-736. |
| [58] | Makarova KS, Zhang F, Koonin EV. SnapShot: class 2 CRISPR-cas systems [J]. Cell, 2017, 168(1/2): 328-328.e1. |
| [59] | Cho S, Shin J, Cho BK. Applications of CRISPR/cas system to bacterial metabolic engineering [J]. Int J Mol Sci, 2018, 19(4): 1089. |
| [60] | Azam MW, Khan AU. CRISPRi-mediated suppression of E. coli Nissle 1917 virulence factors: a strategy for creating an engineered probiotic using csgD gene suppression [J]. Front Nutr, 2022, 9: 938989. |
| [61] | Xu J, Xia K, Li PY, et al. Functional investigation of the chromosomal ccdAB and hipAB operon in Escherichia coli Nissle 1917 [J]. Appl Microbiol Biotechnol, 2020, 104(15): 6731-6747. |
| [62] | Neil K, Allard N, Roy P, et al. High-efficiency delivery of CRISPR-Cas9 by engineered probiotics enables precise microbiome editing [J]. Mol Syst Biol, 2021, 17(10): e10335. |
| [63] | World Health O. The evolving threat of antimicrobial resistance : options for action [M]. Geneva: World Health Organization. 2012. |
| [64] | Bikard D, Euler CW, Jiang WY, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials [J]. Nat Biotechnol, 2014, 32(11): 1146-1150. |
| [65] | Reuter A, Hilpert C, Dedieu-Berne A, et al. Targeted-antibacterial-plasmids (TAPs) combining conjugation and CRISPR/Cas systems achieve strain-specific antibacterial activity [J]. Nucleic Acids Res, 2021, 49(6): 3584-3598. |
| [66] | Rottinghaus AG, Ferreiro A, Fishbein SRS, et al. Genetically stable CRISPR-based kill switches for engineered microbes [J]. Nat Commun, 2022, 13: 672. |
| [67] | Hwang IY, Koh E, Wong A, et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models [J]. Nat Commun, 2017, 8: 15028. |
| [68] | Chan CTY, Lee JW, Cameron DE, et al. ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment [J]. Nat Chem Biol, 2016, 12(2): 82-86. |
| [69] | Din MO, Danino T, Prindle A, et al. Synchronized cycles of bacterial lysis for in vivo delivery [J]. Nature, 2016, 536(7614): 81-85. |
| [70] | Gurbatri CR, Lia I, Vincent R, et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies [J]. Sci Transl Med, 2020, 12(530): eaax0876. |
| [71] | Blum-Oehler G, Oswald S, Eiteljörge K, et al. Development of strain-specific PCR reactions for the detection of the probiotic Escherichia coli strain Nissle 1917 in fecal samples [J]. Res Microbiol, 2003, 154(1): 59-66. |
| [72] | Kan A, Gelfat I, Emani S, et al. Plasmid vectors for in vivo selection-free use with the probiotic E. coli Nissle 1917 [J]. ACS Synth Biol, 2021, 10(1): 94-106. |
| [73] | Buddenborg C, Daudel D, Liebrecht S, et al. Development of a tripartite vector system for live oral immunization using a Gram-negative probiotic carrier [J]. Int J Med Microbiol, 2008, 298(1/2): 105-114. |
| [74] | Whelan RA, Rausch S, Ebner F, et al. A transgenic probiotic secreting a parasite immunomodulator for site-directed treatment of gut inflammation [J]. Mol Ther, 2014, 22(10): 1730-1740. |
| [75] | Ou BM, Jiang BY, Jin D, et al. Engineered recombinant Escherichia coli probiotic strains integrated with F4 and F18 fimbriae cluster genes in the chromosome and their assessment of immunogenic efficacy in vivo [J]. ACS Synth Biol, 2020, 9(2): 412-426. |
| [76] | Lynch JP, Goers L, Lesser CF. Emerging strategies for engineering Escherichia coli Nissle 1917-based therapeutics [J]. Trends Pharmacol Sci, 2022, 43(9): 772-786. |
| [77] | Michael H, Paim FC, Miyazaki A, et al. Escherichia coli Nissle 1917 administered as a dextranomar microsphere biofilm enhances immune responses against human rotavirus in a neonatal malnourished pig model colonized with human infant fecal microbiota [J]. PLoS One, 2021, 16(2): e0246193. |
| [78] | Michael H, Srivastava V, Deblais L, et al. The combined Escherichia coli nissle 1917 and tryptophan treatment modulates immune and metabolome responses to human rotavirus infection in a human infant fecal microbiota-transplanted malnourished gnotobiotic pig model [J]. mSphere, 2022, 7(5) |
| [79] | Sarate PJ, Srutkova D, Geissler N, et al. Pre- and neonatal imprinting on immunological homeostasis and epithelial barrier integrity by Escherichia coli Nissle 1917 prevents allergic poly-sensitization in mice [J]. Front Immunol, 2021, 11: 612775. |
| [80] | Khan I, Ullah N, Zha LJ, et al. Alteration of gut microbiota in inflammatory bowel disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome [J]. Pathogens, 2019, 8(3): 126. |
| [81] | Wang SB, Ghosh AK, Bongio N, et al. Fighting malaria with engineered symbiotic bacteria from vector mosquitoes [J]. Proc Natl Acad Sci USA, 2012, 109(31): 12734-12739. |
| [82] | Wang LF, Liao Y, Yang RB, et al. An engineered probiotic secreting Sj16 ameliorates colitis via Ruminococcaceae/butyrate/retinoic acid axis [J]. Bioeng Transl Med, 2021, 6(3): e10219. |
| [83] | Wang LF, Xie H, Xu L, et al. rSj16 protects against DSS-induced colitis by inhibiting the PPAR-α signaling pathway [J]. Theranostics, 2017, 7(14): 3446-3460. |
| [84] | Baidoun F, Elshiwy K, Elkeraie Y, et al. Colorectal cancer epidemiology: recent trends and impact on outcomes [J]. Curr Drug Targets, 2021, 22(9): 998-1009. |
| [85] | Chen JH, Li XH, Liu YM, et al. Engineering a probiotic strain of Escherichia coli to induce the regression of colorectal cancer through production of 5-aminolevulinic acid [J]. Microb Biotechnol, 2021, 14(5): 2130-2139. |
| [86] | Dougherty TJ, Gomer CJ, Henderson BW, et al. Photodynamic therapy [J]. JNCI J Natl Cancer Inst, 1998, 90(12): 889-905. |
| [87] | Fujino M, Nishio Y, Ito H, et al. 5-Aminolevulinic acid regulates the inflammatory response and alloimmune reaction [J]. Int Immunopharmacol, 2016, 37: 71-78. |
| [88] | Kim CH, Chung CW, Lee HM, et al. Synergistic effects of 5-aminolevulinic acid based photodynamic therapy and celecoxib via oxidative stress in human cholangiocarcinoma cells [J]. Int J Nanomedicine, 2013, 8: 2173-2186. |
| [89] | Isabella VM, Ha BN, Castillo MJ, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria [J]. Nat Biotechnol, 2018, 36(9): 857-864. |
| [90] | Puurunen MK, Vockley J, Searle SL, et al. Safety and pharmacodynamics of an engineered E. coli Nissle for the treatment of phenylketonuria: a first-in-human phase 1/2a study [J]. Nat Metab, 2021, 3(8): 1125-1132. |
| [91] | Jiang Y, Sun BB, Qian FH, et al. Expression of phenylalanine ammonia lyase as an intracellularly free and extracellularly cell surface-immobilized enzyme on a gut microbe as a live biotherapeutic for phenylketonuria [J]. Sci China Life Sci, 2023, 66(1): 127-136. |
| [92] | Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022 [J]. CA A Cancer J Clin, 2022, 72(1): 7-33. |
| [93] | Gustafson KT, Huynh KT, Heineck D, et al. Automated fluorescence quantification of extracellular vesicles collected from blood plasma using dielectrophoresis [J]. Lab Chip, 2021, 21(7): 1318-1332. |
| [94] | Hinestrosa JP, Searson DJ, Lewis JM, et al. Simultaneous isolation of circulating nucleic acids and EV-associated protein biomarkers from unprocessed plasma using an AC electrokinetics-based platform [J]. Front Bioeng Biotechnol, 2020, 8: 581157. |
| [95] | Ibsen S, Sonnenberg A, Schutt C, et al. Recovery of drug delivery nanoparticles from human plasma using an electrokinetic platform technology [J]. Small, 2015, 11(38): 5088-5096. |
| [96] | Geervliet M, Lute LCP, Jansen CA, et al. Differential immunomodulation of porcine bone marrow derived dendritic cells by E. coli Nissle 1917 and β-glucans [J]. PLoS One, 2020, 15(6): e0233773. |
| [97] | Secher T, Maillet I, Mackowiak C, et al. The probiotic strain Escherichia coli Nissle 1917 prevents papain-induced respiratory barrier injury and severe allergic inflammation in mice [J]. Sci Rep, 2018, 8: 11245. |
| [98] | Bury S, Soundararajan M, Bharti R, et al. The probiotic Escherichia coli strain Nissle 1917 combats lambdoid bacteriophages stx and λ [J]. Front Microbiol, 2018, 9: 929. |
| [99] | Helmy YA, Kassem II, et al. Immuno-modulatory effect of probiotic E. coli Nissle 1917 in polarized human colonic cells against Campylobacter jejuni infection [J]. Gut Microbes, 2021, 13: 1857514. |
| [100] | Dusek O, Fajstova A, Klimova A, et al. Severity of experimental autoimmune uveitis is reduced by pretreatment with live probiotic Escherichia coli Nissle 1917 [J]. Cells, 2020, 10(1): 23. |
| [101] | Ji JJ, Sun QM, Nie DY, et al. Probiotics protect against RSV infection by modulating the microbiota-alveolar-macrophage axis [J]. Acta Pharmacol Sin, 2021, 42(10): 1630-1641. |
| [102] | Park K, Park S, Nagappan A, et al. Probiotic Escherichia coli ameliorates antibiotic-associated anxiety responses in mice [J]. Nutrients, 2021, 13(3): 811. |
| [103] | Manfredi C, Calace FP, Fusco F, et al. Escherichia coli Nissle 1917 as adjuvant therapy in patients with chronic bacterial prostatitis: a non-blinded, randomized, controlled trial [J]. World J Urol, 2021, 39(12): 4373-4379. |
| [104] | Hu RJ, Lin H, Li J, et al. Probiotic Escherichia coli Nissle 1917-derived outer membrane vesicles enhance immunomodulation and antimicrobial activity in RAW264.7 macrophages [J]. BMC Microbiol, 2020, 20(1): 268. |
| [105] | Hamilton S, Shea D, Ibsen S, et al. On-chip dielectrophoretic recovery and detection of a lactate sensing probiotic from model human saliva [J]. Electrophoresis, 2023, 44(3/4): 442-449. |
| [106] | Droste JHJ, Wieringa MH, Weyler JJ, et al. Does the use of antibiotics in early childhood increase the risk of asthma and allergic disease? [J]. Clin Exp Allergy, 2000, 30(11): 1548-1553. |
| [107] | Gupta RS, Singh AM, Walkner M, et al. Hygiene factors associated with childhood food allergy and asthma [J]. Allergy Asthma Proc, 2016, 37(6): 140-146. |
| [108] | Geldart KG, Kommineni S, Forbes M, et al. Engineered E. coli Nissle 1917 for the reduction of vancomycin-resistant Enterococcus in the intestinal tract [J]. Bioeng Transl Med, 2018, 3(3): 197-208. |
| [109] | Forkus B, Ritter S, Vlysidis M, et al. Antimicrobial probiotics reduce Salmonella enterica in Turkey gastrointestinal tracts [J]. Sci Rep, 2017, 7: 40695. |
| [110] | Wang LL, Mao YF, Wang ZW, et al. Advances in biotechnological production of β-alanine [J]. World J Microbiol Biotechnol, 2021, 37(5): 79. |
| [111] | Wang P, Zhou HY, Li B, et al. Multiplex modification of Escherichia coli for enhanced β-alanine biosynthesis through metabolic engineering [J]. Bioresour Technol, 2021, 342: 126050. |
| [112] | Dong YF, Chen Z. Systems metabolic engineering of Corynebacterium glutamicum for efficient L-tryptophan production [J]. Synth Syst Biotechnol, 2025, 10(2): 511-522. |
| [113] | Hu SL, Fei MY, Fu BB, et al. Development of probiotic E. coli Nissle 1917 for β-alanine production by using protein and metabolic engineering [J]. Appl Microbiol Biotechnol, 2023, 107(7): 2277-2288. |
| [114] | Datta P, Fu L, Brodfuerer P, et al. High density fermentation of probiotic E. coli Nissle 1917 towards heparosan production, characterization, and modification [J]. Appl Microbiol Biotechnol, 2021, 105(3): 1051-1062. |
| [115] | Datta P, Yan LF, Awofiranye A, et al. Heparosan chain characterization: sequential depolymerization of E. coli K5 heparosan by a bacterial eliminase heparin lyase III and a bacterial hydrolase heparanase bp to prepare defined oligomers [J]. Biotechnol J, 2021, 16(3): 2000336. |
| [116] | Hu S, Zhou SY, Wang Y, et al. Coordinated optimization of the polymerization and transportation processes to enhance the yield of exopolysaccharide heparosan [J]. Carbohydr Polym, 2024, 333: 121983. |
| [117] | Sajadi E, Fatemi SS, Babaeipour V, et al. Increased cellulose production by heterologous expression of bcsA and B genes from Gluconacetobacterxylinus in E. coli Nissle 1917 [J]. Bioprocess Biosyst Eng, 2019, 42(12): 2023-2034. |
| [118] | Swanson D, Block R, Mousa SA. Omega-3 fatty acids EPA and DHA: health benefits throughout life [J]. Adv Nutr, 2012, 3(1): 1-7. |
| [119] | Amiri-Jami M, Abdelhamid AG, Hazaa M, et al. Recombinant production of omega-3 fatty acids by probiotic Escherichia coli Nissle 1917 [J]. FEMS Microbiol Lett, 2015, 362(20): fnv166. |
| [120] | Busch AWU, Montgomery BL. Interdependence of tetrapyrrole metabolism, the generation of oxidative stress and the mitigative oxidative stress response [J]. Redox Biol, 2015, 4: 260-271. |
| [121] | Canesin G, Hejazi SM, Swanson KD, et al. Heme-derived metabolic signals dictate immune responses [J]. Front Immunol, 2020, 11: 66. |
| [122] | Rochette L, Zeller M, Cottin Y, et al. Redox functions of heme oxygenase-1 and biliverdin reductase in diabetes [J]. Trends Endocrinol Metab, 2018, 29(2): 74-85. |
| [123] | Kurtz CB, Millet YA, Puurunen MK, et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans [J]. Sci Transl Med, 2019, 11(475): eaau7975. |
| [124] | Park YT, Kim T, Ham J, et al. Physiological activity of E. coli engineered to produce butyric acid [J]. Microb Biotechnol, 2022, 15(3): 832-843. |
| [125] | Chiang CJ, Hong YH. In situ delivery of biobutyrate by probiotic Escherichia coli for cancer therapy [J]. Sci Rep, 2021, 11: 18172. |
| [126] | Ma J, Li CY, Wang JR, et al. Genetically engineered Escherichia coli Nissle 1917 secreting GLP-1 analog exhibits potential antiobesity effect in high-fat diet-induced obesity mice [J]. Obesity, 2020, 28(2): 315-322. |
| [127] | Kotopka BJ, Smolke CD. Model-driven generation of artificial yeast promoters [J]. Nat Commun, 2020, 11: 2113. |
| [128] | Van Brempt M, Clauwaert J, Mey F, et al. Predictive design of sigma factor-specific promoters [J]. Nat Commun, 2020, 11: 5822. |
| [129] | Zhao M, Yuan ZQ, Wu LT, et al. Precise prediction of promoter strength based on a de novo synthetic promoter library coupled with machine learning [J]. ACS Synth Biol, 2022, 11(1): 92-102. |
| [130] | Jervis A, Carbonell P, Vinaixa M, et al. Machine learning of designed translational control allows predictive pathway optimization in Escherichia coli [J]. ACS Synth Biol, 2019, 8(1): 127-136. |
| [131] | Meng HL, Wang JF, Xiong ZQ, et al. Quantitative design of regulatory elements based on high-precision strength prediction using artificial neural network [J]. PLoS One, 2013, 8(4): e60288. |
| [132] | Chari R, Yeo NC, Chavez A, et al. sgRNA scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity [J]. ACS Synth Biol, 2017, 6(5): 902-904. |
| [133] | Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9 [J]. Nat Biotechnol, 2016, 34(2): 184-191. |
| [134] | Kim HK, Min S, Song M, et al. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity [J]. Nat Biotechnol, 2018, 36(3): 239-241. |
| [135] | Yu TH, Cui HY, Li JC, et al. Enzyme function prediction using contrastive learning [J]. Science, 2023, 379(6639): 1358-1363. |
| [136] | Wu Z, Jennifer Kan SB, Lewis RD, et al. Machine learning-assisted directed protein evolution with combinatorial libraries [J]. Proc Natl Acad Sci U S A, 2019, 116(18): 8852-8858. |
| [137] | Fox RJ, Davis SC, Mundorff EC, et al. Improving catalytic function by ProSAR-driven enzyme evolution [J]. Nat Biotechnol, 2007, 25(3): 338-344. |
| [138] | Saito Y, Oikawa M, Nakazawa H, et al. Machine-learning-guided mutagenesis for directed evolution of fluorescent proteins [J]. ACS Synth Biol, 2018, 7(9): 2014-2022. |
| [139] | Romero PA, Krause A, Arnold FH. Navigating the protein fitness landscape with Gaussian processes [J]. Proc Natl Acad Sci U S A, 2013, 110(3): E193-E201. |
| [140] | Radivojević T, Costello Z, Workman K, et al. A machine learning Automated Recommendation Tool for synthetic biology [J]. Nat Commun, 2020, 11: 4879. |
| [141] | Christopher Culley SV. A mechanism-aware and multiomic machine-learning pipeline characterizes yeast cell growth [J]. Proc Natl Acad Sci U S A, 2020, 117(31): 18869-18879. |
| [142] | Vijayakumar S, Rahman PKSM, Angione C. A hybrid flux balance analysis and machine learning pipeline elucidates metabolic adaptation in cyanobacteria [J]. iScience, 2020, 23(12): 101818. |
| [143] | Sample PJ, Wang B, Reid DW, et al. Human 5' UTR design and variant effect prediction from a massively parallel translation assay [J]. Nat Biotechnol, 2019, 37(7): 803-809. |
| [144] | Clauwaert J, Menschaert G, Waegeman W. DeepRibo: a neural network for precise gene annotation of prokaryotes by combining ribosome profiling signal and binding site patterns [J]. Nucleic Acids Res, 2019, 47(6): e36. |
| [145] | Ryu JY, Kim HU, Lee SY. Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers [J]. Proc Natl Acad Sci U S A, 2019, 116(28): 13996-14001. |
| [146] | Senior AW, Evans R, Jumper J, et al. Improved protein structure prediction using potentials from deep learning [J]. Nature, 2020, 577(7792): 706-710. |
| [147] | Stokes JM, Yang K, Swanson K, et al. A deep learning approach to antibiotic discovery [J]. Cell, 2020, 181(2): 475-483. |
| [148] | Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation [M]//Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015. Cham: Springer International Publishing, 2015: 234-241. |
| [149] | Treloar NJ, Fedorec AJH, Ingalls B, et al. Deep reinforcement learning for the control of microbial co-cultures in bioreactors [J]. PLoS Comput Biol, 2020, 16(4): e1007783. |
| [150] | Sarkar K, Bonnerjee D, Srivastava R, et al. A single layer artificial neural network type architecture with molecular engineered bacteria for reversible and irreversible computing [J]. Chem Sci, 2021, 12(48): 15821-15832. |
| [151] | Li XM, Rizik L, Kravchik V, et al. Synthetic neural-like computing in microbial consortia for pattern recognition [J]. Nat Commun, 2021, 12: 3139. |
| [152] | Adikusuma F, Piltz S, Corbett MA, et al. Large deletions induced by Cas9 cleavage [J]. Nature, 2018, 560(7717): E8-E9. |
| [153] | Cullot G, Boutin J, Toutain J, et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations [J]. Nat Commun, 2019, 10: 1136. |
| [154] | Selle K, Fletcher JR, Tuson H, et al. In vivo targeting of Clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials [J]. mBio, 2020, 11(2) |
| [1] | WANG Cong-huan, WU Guo-qiang, WEI Ming. Functional Mechanism of Plant CBL in Regulating the Responses to Abiotic and Biotic Stresses [J]. Biotechnology Bulletin, 2025, 41(7): 1-16. |
| [2] | WANG Xin, SUN Tao, SUN Mei-li, WANG Kai-feng, JI Xiao-jun. Advances in the Biosynthesis of Functional Food Ingredient Hydroxytyrosol [J]. Biotechnology Bulletin, 2025, 41(11): 35-46. |
| [3] | HU Miao-miao, QIN Shu-fang. Application and Research Progress of Synthetic Biology in the Synthesis of Human Milk Oligosaccharides [J]. Biotechnology Bulletin, 2025, 41(11): 28-34. |
| [4] | SUN Jing, YANG Yun-long, LIU Rong-zhi, YU Hong, LU Tie-gang. Strengthening Fundamental Research on Photosynthesis to Support Crop Breeding for High Yield [J]. Biotechnology Bulletin, 2025, 41(10): 1-5. |
| [5] | GAO Bo-wen, DING Shun-hua, CHEN Xiao-jun, WEN Xiao-gang, TIAN Li-jin, LU Qing-tao. Strategies for Optimizing Photosynthesis to Enhance Agricultural Production Efficiency [J]. Biotechnology Bulletin, 2025, 41(10): 54-63. |
| [6] | WANG Zi-rui, LIU Xiao-han, XIU Yu, LIN Shan-zhi. Cloning of the Promoter of MATE40 Gene from Prunus sibirica Seeds and Analysis of Gene Expression Driven by Promoter [J]. Biotechnology Bulletin, 2024, 40(11): 192-201. |
| [7] | JU Kang-hui, TIAN Xiao-ya, WANG Li, CHEN Jing-yu. Cell Programming Technology: Paving the Way for Efficient Cell Factories [J]. Biotechnology Bulletin, 2024, 40(10): 160-171. |
| [8] | CHENG Ting, YUAN Shuai, ZHANG Xiao-yuan, LIN Liang-cai, LI Xin, ZHANG Cui-ying. Research Progress in the Regulation of Isobutanol Synthesis Pathway in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(7): 80-90. |
| [9] | CHEN Cai-ping, REN Hao, LONG Teng-fei, HE Bing, LU Zhao-xiang, SUN Jian. Research Advances in the Treatment of Inflammation Bowel Disease Using Escherichia coli Nissle 1917 [J]. Biotechnology Bulletin, 2023, 39(6): 109-118. |
| [10] | WANG Xiao-mei, YANG Xiao-wei, LI Hui-shang, HE Wei, XIN Zhu-lin. Development Status of Synthetic Biology in Globe and Its Enlightenment [J]. Biotechnology Bulletin, 2023, 39(2): 292-302. |
| [11] | CHEN Xiao-lin, LIU Yang-er, XU Wen-tao, GUO Ming-zhang, LIU Hui-lin. Application of Synthetic Biology Based Whole-cell Biosensor Technology in the Rapid Detection of Food Safety [J]. Biotechnology Bulletin, 2023, 39(1): 137-149. |
| [12] | ZHOU Lin, LIANG Xuan-ming, ZHAO Lei. Biosynthesis of Natural Carotenoids:Progress and Perspective [J]. Biotechnology Bulletin, 2022, 38(7): 119-127. |
| [13] | ZHONG Ming-yue, LIU Chun-yan, YAN Yan, ZHANG Xiao-hui, YUAN Hai-sheng, XU Guo-quan, ZHANG He-ping, WANG Yu-zhen. Improvement Effect of Bifidobacterium lactis V9 on NAFLD Rats Induced by High-fat Diet [J]. Biotechnology Bulletin, 2022, 38(3): 181-187. |
| [14] | GUO Xiao-zhen, ZHANG Xue-fu. Analysis of the Development Trend in the Field of Plant Synthetic Biology [J]. Biotechnology Bulletin, 2022, 38(2): 289-296. |
| [15] | ZHAO Yu-xue, WANG Yun, YU Lu-yao, LIU Jing-jing, SI Jin-ping, ZHANG Xin-feng, ZHANG Lei. Structure and Application of C-glycosyltransferases in Plants [J]. Biotechnology Bulletin, 2022, 38(10): 18-28. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||