








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (12): 191-197.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0230
Previous Articles Next Articles
FAN Yu-chen( ), LU Yao, LIU Xiang-nan, ZHAO Bo(
), LU Yao, LIU Xiang-nan, ZHAO Bo( )
)
Received:2021-03-01
Online:2021-12-26
Published:2022-01-19
Contact:
ZHAO Bo
E-mail:sjtufyc@sjtu.edu.cn;bozhao@sjtu.edu.cn
FAN Yu-chen, LU Yao, LIU Xiang-nan, ZHAO Bo. Construction of Mutants Swapping Ubiquitin E3 Ligase CHIP and E4B U-box Domain and Verification of Ubiquitination Activity[J]. Biotechnology Bulletin, 2021, 37(12): 191-197.
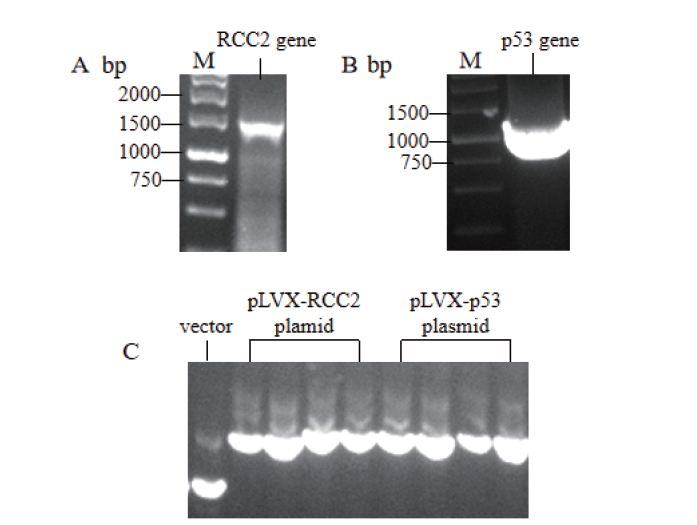
Fig.1 Plasmid cloning and construction of substrate RCC2 and p53 M:DNA molecular weight standard. A:PCR products of RCC2 gene. B:PCR products of p53 gene. C:Electrophoresis band of pLVX empty vector plasmid and positive cloned plasmid
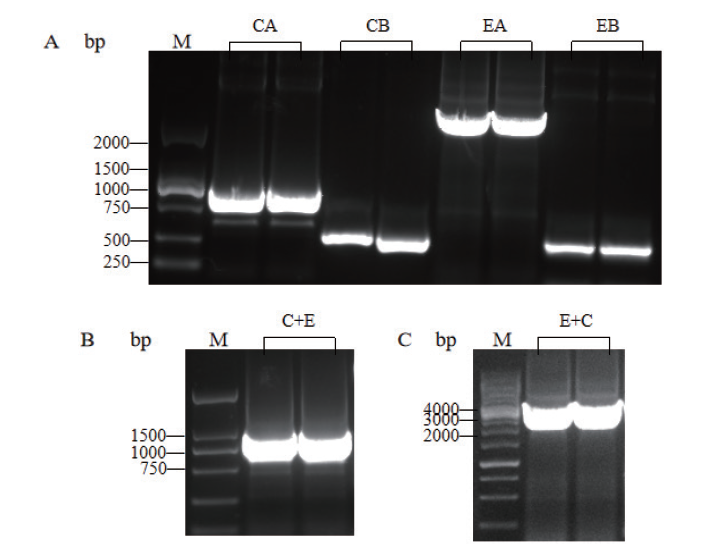
Fig.2 PCR results of fusion genes C+E and E+C M:DNA molecular weight standard. A:PCR products of CA,CB,EA and EB fragments. B:PCR products of C+E gene. C:PCR products of E+C gene
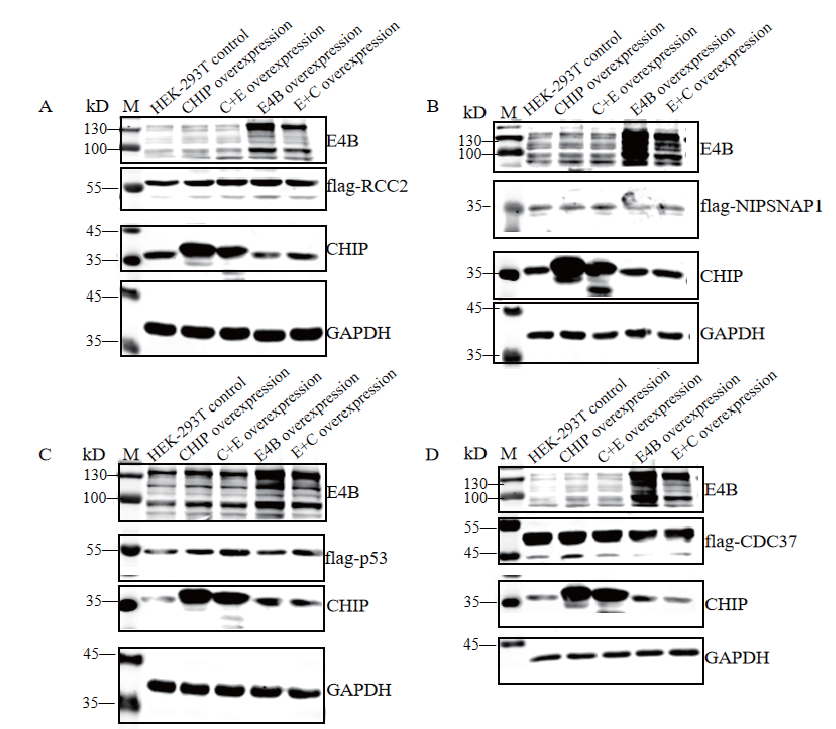
Fig.3 Intracellular expression results of substrates,CHIP,E4B and their mutants M:Protein molecular weight standard. A,B,C and D:Expression levels of substrates,CHIP,E4B and their mutants in cell lysates
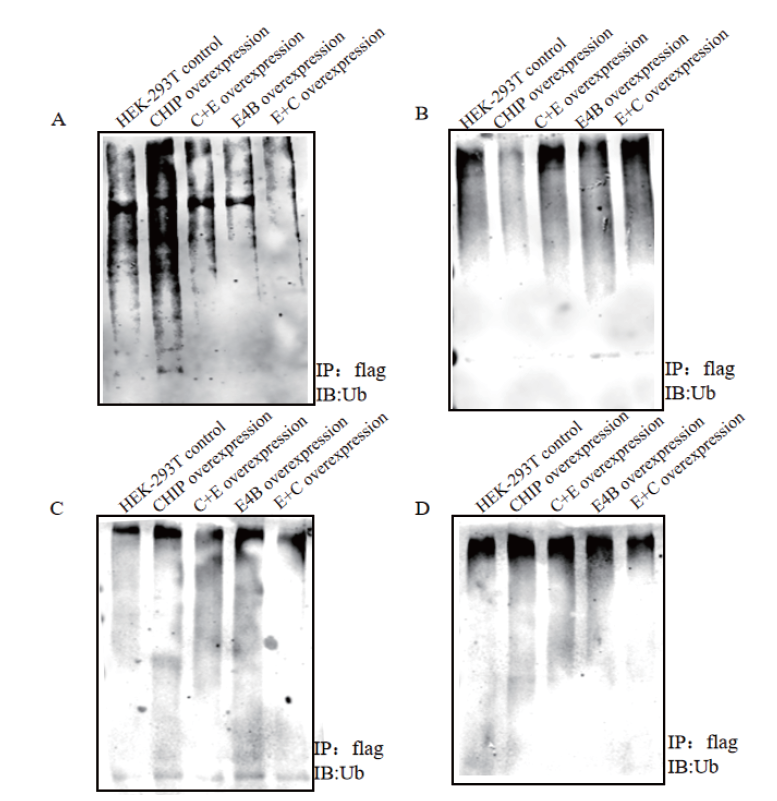
Fig.4 Co-immunoprecipitation experiment results of sub-strate RCC2,NIPSNAP1,p53 and CDC37 A,B,C and D:Detection of the ubiquitination activity of CHIP,E4B and their mutants on the substrates in the cell by co-immunoprecipitation
| [1] |
Hu H, Sun SC. Ubiquitin signaling in immune responses[J]. Cell Res, 2016, 26(4):457-483.
doi: 10.1038/cr.2016.40 URL |
| [2] |
Sharma B, Taganna J. Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato[J]. Sci Rep, 2020, 10(1):9581.
doi: 10.1038/s41598-020-66553-1 URL |
| [3] |
Pruneda JN, Littlefield PJ, Soss SE, et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases[J]. Mol Cell, 2012, 47(6):933-942.
doi: 10.1016/j.molcel.2012.07.001 URL |
| [4] |
Hatakeyama S, Matsumoto M, Yada M, et al. Interaction of U-box-type ubiquitin-protein ligases(E3s)with molecular chaperones[J]. Genes Cells, 2004, 9(6):533-548.
doi: 10.1111/gtc.2004.9.issue-6 URL |
| [5] |
Nikolay R, Wiederkehr T, Rist W, et al. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity[J]. J Biol Chem, 2004, 279(4):2673-2678.
pmid: 14610072 |
| [6] |
Susaki E, Kaneko-Oshikawa C, Miyata K, et al. Increased E4 activity in mice leads to ubiquitin-containing aggregates and degeneration of hypothalamic neurons resulting in obesity[J]. J Biol Chem, 2010, 285(20):15538-15547.
doi: 10.1074/jbc.M110.105841 pmid: 20190229 |
| [7] |
Zhang P, Li C, Li H, et al. Ubiquitin ligase CHIP regulates OTUD3 stability and suppresses tumour metastasis in lung cancer[J]. Cell Death Differ, 2020, 27(11):3177-3195.
doi: 10.1038/s41418-020-0571-7 URL |
| [8] |
Starita LM, Pruneda JN, Lo RS, et al. Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis[J]. PNAS, 2013, 110(14):E1263-E1272.
doi: 10.1073/pnas.1303309110 URL |
| [9] |
Zhong D, Ru Y, Wang Q, et al. Chimeric ubiquitin ligases inhibit non-small cell lung cancer via negative modulation of EGFR signaling[J]. Cancer Lett, 2015, 359(1):57-64.
doi: 10.1016/j.canlet.2014.12.043 URL |
| [10] |
Bhuripanyo K, Wang Y, Liu X, et al. Identifying the substrate proteins of U-box E3s E4B and CHIP by orthogonal ubiquitin transfer[J]. Sci Adv, 2018, 4(1):e1701393.
doi: 10.1126/sciadv.1701393 URL |
| [11] |
Wu H, Pomeroy SL, Ferreira M, et al. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53[J]. Nat Med, 2011, 17(3):347-355.
doi: 10.1038/nm.2283 URL |
| [12] |
Meyer-Schwesinger C. The ubiquitin-proteasome system in kidney physiology and disease[J]. Nat Rev Nephrol, 2019, 15(7):393-411.
doi: 10.1038/s41581-019-0148-1 pmid: 31036905 |
| [13] |
Yoo L, Yoon AR, Yun CO, et al. Covalent ISG15 conjugation to CHIP promotes its ubiquitin E3 ligase activity and inhibits lung cancer cell growth in response to type I interferon[J]. Cell Death Dis, 2018, 9(2):97.
doi: 10.1038/s41419-017-0138-9 URL |
| [14] |
Xu J, Wang H, Li W, et al. E3 ubiquitin ligase CHIP attenuates cellular proliferation and invasion abilities in triple-negative breast cancer cells[J]. Clin Exp Med, 2020, 20(1):109-119.
doi: 10.1007/s10238-019-00594-3 URL |
| [15] |
Madrigal SC, McNeil Z, Sanchez-Hodge R, et al. Changes in protein function underlie the disease spectrum in patients with CHIP mutations[J]. J Biol Chem, 2019, 294(50):19236-19245.
doi: 10.1074/jbc.RA119.011173 pmid: 31619515 |
| [16] |
Ru Y, Wang Q, Liu X, et al. The chimeric ubiquitin ligase SH2-U-box inhibits the growth of imatinib-sensitive and resistant CML by targeting the native and T315I-mutant BCR-ABL[J]. Sci Rep, 2016, 6:28352.
doi: 10.1038/srep28352 URL |
| [17] | 陆瑶, 赵博, 武正华. 泛素连接酶UBE4B的研究进展[J]. 生物学杂志, 2020, 37(5):90-93. |
| Lu Y, Zhao B, Wu ZH. The progress on the ubiquitin ligase UBE4B[J]. J Biol, 2020, 37(5):90-93. | |
| [18] |
Zhang Y, Lv Y, Zhang Y, et al. Regulation of p53 level by UBE4B in breast cancer[J]. PLoS One, 2014, 9(2):e90154.
doi: 10.1371/journal.pone.0090154 URL |
| [19] |
Hellerschmied D, Roessler M, Lehner A, et al. UFD-2 is an adaptor-assisted E3 ligase targeting unfolded proteins[J]. Nat Commun, 2018, 9(1):484.
doi: 10.1038/s41467-018-02924-7 pmid: 29396393 |
| [1] | LI Wang-ning, ZHANG Hao-jie, LI Ya-nan, LIANG Meng-jing, JI Chun-li, Zhang Chun-hui, LI Run-zhi, CUI Yu-lin, QIN Song, CUI Hong-li. Phenotypic Characterization of Blue Photoreceptor Plant Type Cryptochrome CRY Mutant in Chlamydomonas reinhardtii [J]. Biotechnology Bulletin, 2023, 39(2): 243-253. |
| [2] | CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. Identification and Gene Functional Analysis of Salinity-hypersensitive Mutant ss2 in Rice [J]. Biotechnology Bulletin, 2022, 38(9): 158-166. |
| [3] | TANG Guang-fu, GUI Yan-ling, MAN Hai-qiao, ZHAO Jie-hong. Editing pyrG Gene of Monascus by CRISPR/Cas 9 and Its Effects on Secondary Metabolism [J]. Biotechnology Bulletin, 2022, 38(8): 198-205. |
| [4] | ZHOU Shi-chen, YI Zhi-ben, WANG Xin-yi, YANG Xiao-ying, SUN Li-na, LUAN Wei-jiang, LIANG Shan-shan. Genetic Analysis and Gene Mapping of Sorghum Double-grain Mutant Dgs [J]. Biotechnology Bulletin, 2022, 38(7): 171-177. |
| [5] | TANG Yue-hui, ZHAO Yu-fan, LIN Jin, WANG Yin, CAO Bo-yuan, CHE Yi-fan, YANG Wen-jie, BAO Xin-xin, YANG Tong-wen. Identification and Gene Mapping of a Seedling Lethal Mutant in Rice [J]. Biotechnology Bulletin, 2022, 38(10): 124-131. |
| [6] | XIONG Xue, LI Peng, ZHANG Gui-he, XIANG Zhun, TAO Wen-Guang, ZHOU Guang-yan, HE Yao-wei. Effects of Different Cultivation Substrates on the Laccase Activities of Lentinula edodes During Liquid Fermentation [J]. Biotechnology Bulletin, 2021, 37(12): 50-59. |
| [7] | TAO Zhi-dong, HE Yan-hui, DENG Zi-he, SUN Lin-lin, WU Zhan-sheng. Screening of High-efficiency Cellulose-degrading Microorganism from Spent Lentinula edodes Substrate and Optimization of Its Enzyme Production [J]. Biotechnology Bulletin, 2021, 37(11): 158-165. |
| [8] | ZHU Cai-lin, LÜ Xiang, XIA Xiao-le. Effect of Site-directed Mutagenesis of Amino Acids in Lid Region on the Enzymatic Properties of T1 Lipase [J]. Biotechnology Bulletin, 2020, 36(11): 94-102. |
| [9] | CAI Yu-zhen, BAI Qiao-yan, SU Min, TANG Liang-hua. Strategies and Advances in the Molecular Modification of Substrate Binding Pocket of Lipase [J]. Biotechnology Bulletin, 2020, 36(11): 173-180. |
| [10] | BI Yan-zhen, XIAO Hong-wei, ZHANG Li-ping, REN Hong-yan, HUA Zai-dong, HUA Wen-jun, WANG Zheng, NIU Min-jie, LIN Zheng-yun, REN Xi-dong, SUN Li-hua, ZHENG Xin-min. Applications and Challenges of Gene Editing in Non-human Primate Models [J]. Biotechnology Bulletin, 2018, 34(5): 48-56. |
| [11] | LI Su-zhen, YANG Wen-zhu, CHEN Ru-mei. An Overview on Yellow Green Leaf Mutants in Rice [J]. Biotechnology Bulletin, 2018, 34(11): 15-21. |
| [12] | WANG Wen-xiu, WANG Lei. Research Progress on Maize Dwarf Genes [J]. Biotechnology Bulletin, 2018, 34(11): 22-26. |
| [13] | LIU Qi-ming,CHEN Zhen-hua,HAN Xiao. Monosaccharide Components of the Male Sterile Mutant gsl5 in Rice [J]. Biotechnology Bulletin, 2017, 33(9): 116-119. |
| [14] | WANG Jia-mei, AN Xue-jiao, ZHANG Zhi-guo. Mapping-based Cloning of a Wax Crystal-Sparse Leaf Mutant wcl1 in Rice [J]. Biotechnology Bulletin, 2017, 33(8): 46-50. |
| [15] | LIU Xiao-li, JIANG Shi-jie, XUE Dong, LIU Ying-ying, WU Xiao-li, FENG Shuai, HAN Jia-hui, WANG Yu-zhou, PING Shu-zhen, WANG Jin. Construction and Biological Characterization of Gene dlp Deletion Mutant of Deinococcus radiodurans R1 [J]. Biotechnology Bulletin, 2017, 33(2): 155-163. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||