








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (6): 13-23.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1343
Previous Articles Next Articles
HAO Xiang-yang1( ), LIU Fan1, WU Huan1, WANG Bin1, SUN Xue-li1, XIANG Lei-lei1, WANG Tian-chi1, LAI Zhong-xiong1(
), LIU Fan1, WU Huan1, WANG Bin1, SUN Xue-li1, XIANG Lei-lei1, WANG Tian-chi1, LAI Zhong-xiong1( ), CHENG Chun-zhen1,2(
), CHENG Chun-zhen1,2( )
)
Received:2020-11-02
Online:2021-06-26
Published:2021-07-08
Contact:
LAI Zhong-xiong,CHENG Chun-zhen
E-mail:190995955@qq.com;laizx01@163.com;ld0532cheng@126.com
HAO Xiang-yang, LIU Fan, WU Huan, WANG Bin, SUN Xue-li, XIANG Lei-lei, WANG Tian-chi, LAI Zhong-xiong, CHENG Chun-zhen. Cloning and Expression Analysis of GjPAL Genes in Gerbera jamesonni[J]. Biotechnology Bulletin, 2021, 37(6): 13-23.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 产物大小Product size/bp | 用途Application |
|---|---|---|---|
| GjPAL1F-outer | GGATTACGGGTTCAAAGGT | 3' RACE | |
| GjPAL1F-inner | GCTCCAGTTTCTTGCTAATCC | ||
| GjPAL4F-outer | TAAGCCAAGTAGCCAAGAAGGT | ||
| GjPAL4F-inner | AGGACTTGCTTCGTGTGGTT | ||
| GjPAL1R-outer | CGGCGTTCAAGAATCTGA | 5'RACE | |
| GjPAL1R-inner | GTGACACCGTAACTATCAGTCCC | ||
| GjPAL1F | GAGTCCAGTGTGTGAAAC | 2 301 | RT-PCR |
| GjPAL1R | AATGGTGAGCCTCTTCCGA | ||
| GjPAL2F | TGGATCATACCAATGGAAATGAC | 2 137 | |
| GjPAL2R | CACTTATGAAATGGGAAGAGGG | ||
| GjPAL3F | ATGGACAGTAAAGCCGTCAAGATC | 2 204 | |
| GjPAL3R | CTAACAAATTGGAAGAGGAACAC | ||
| GjPAL4F | GAGTCTAGTGTGTGAAACTTTGATAGC | 2 117 | |
| GjPAL4R | TTAACATATCGGAAGAGGCTCAC | ||
| GjPAL1-qF | TTCCGAACAGAATCAAGGC | 125 | qRT-PCR |
| GjPAL1-qR | TGACCCTTACACATTGCCG | ||
| GjPAL2-qF | TGCATGAATAGTGATCCATTG | 145 | |
| GjPAL2-qR | GATTCTGACAACTCCACCTGT | ||
| GjPAL3-qF | TCACATAACCAGCCATTTCG | 173 | |
| GjPAL3-qR | ACCATCCGCTTCACTTCGT | ||
| GjPAL4-qF | CGATAATCAATGGAGAACGG | 172 | |
| GjPAL4-qR | TCCACCATCTGTTTCACCTC | ||
| 18sF | TCAAAGCAAGCCTACGCTCT | 125 | |
| 18sR | GCTTTCGCAGTTGTTCGTCT |
Table 1 Information of the primers used for gene cloning and qRT-PCR
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 产物大小Product size/bp | 用途Application |
|---|---|---|---|
| GjPAL1F-outer | GGATTACGGGTTCAAAGGT | 3' RACE | |
| GjPAL1F-inner | GCTCCAGTTTCTTGCTAATCC | ||
| GjPAL4F-outer | TAAGCCAAGTAGCCAAGAAGGT | ||
| GjPAL4F-inner | AGGACTTGCTTCGTGTGGTT | ||
| GjPAL1R-outer | CGGCGTTCAAGAATCTGA | 5'RACE | |
| GjPAL1R-inner | GTGACACCGTAACTATCAGTCCC | ||
| GjPAL1F | GAGTCCAGTGTGTGAAAC | 2 301 | RT-PCR |
| GjPAL1R | AATGGTGAGCCTCTTCCGA | ||
| GjPAL2F | TGGATCATACCAATGGAAATGAC | 2 137 | |
| GjPAL2R | CACTTATGAAATGGGAAGAGGG | ||
| GjPAL3F | ATGGACAGTAAAGCCGTCAAGATC | 2 204 | |
| GjPAL3R | CTAACAAATTGGAAGAGGAACAC | ||
| GjPAL4F | GAGTCTAGTGTGTGAAACTTTGATAGC | 2 117 | |
| GjPAL4R | TTAACATATCGGAAGAGGCTCAC | ||
| GjPAL1-qF | TTCCGAACAGAATCAAGGC | 125 | qRT-PCR |
| GjPAL1-qR | TGACCCTTACACATTGCCG | ||
| GjPAL2-qF | TGCATGAATAGTGATCCATTG | 145 | |
| GjPAL2-qR | GATTCTGACAACTCCACCTGT | ||
| GjPAL3-qF | TCACATAACCAGCCATTTCG | 173 | |
| GjPAL3-qR | ACCATCCGCTTCACTTCGT | ||
| GjPAL4-qF | CGATAATCAATGGAGAACGG | 172 | |
| GjPAL4-qR | TCCACCATCTGTTTCACCTC | ||
| 18sF | TCAAAGCAAGCCTACGCTCT | 125 | |
| 18sR | GCTTTCGCAGTTGTTCGTCT |
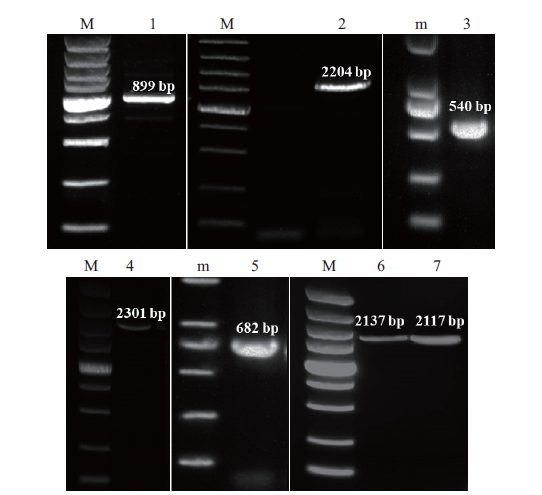
Fig. 1 Electrophoresis result of PCR products 1-7 is the electrophoresis result of GjPAL1 3' RACE, GjPAL3 RT-PCR, GjPAL1 5' RACE, GjPAL1 RT-PCR, GjPAL4 3' RACE, GjPAL2 RT-PCR and GjPAL4 RT-PCR products, respectively. M: DL5000 DNA marker (the band from bottom to top is 100 bp, 250 bp, 500 bp, 750 bp, 1 000 bp, 1 500 bp, 2 000 bp, 3 000 bp and 5 000 bp, respectively). m: DL2000 DNA marker (the band from bottom to top is 100 bp, 250 bp, 500 bp, 750 bp, 1000 bp and 2000 bp, respectively)
| 蛋白 Protein | 分子量 Molecular weight/kD | 分子式 Formula | 等电点pI | 脂肪系数 Aliphatic index | 不稳定系数 Instability index | 总亲水性 GRAVY |
|---|---|---|---|---|---|---|
| GjPAL1 | 77 052.27 | C3405H5452N938O1033S31 | 6.18 | 90.49 | 31.79 | -0.134 |
| GjPAL2 | 76 538.09 | C3384H5389N925O1044S25 | 5.53 | 91.97 | 34.20 | -0.125 |
| GjPAL3 | 77 326.50 | C3428H5470N940O1036S28 | 5.93 | 92.07 | 34.24 | -0.091 |
| GjPAL4 | 77 091.13 | C3406H5453N941O1038S28 | 6.18 | 91.05 | 32.98 | -0.152 |
Table 2 Physicochemical properties of the four GjPALs
| 蛋白 Protein | 分子量 Molecular weight/kD | 分子式 Formula | 等电点pI | 脂肪系数 Aliphatic index | 不稳定系数 Instability index | 总亲水性 GRAVY |
|---|---|---|---|---|---|---|
| GjPAL1 | 77 052.27 | C3405H5452N938O1033S31 | 6.18 | 90.49 | 31.79 | -0.134 |
| GjPAL2 | 76 538.09 | C3384H5389N925O1044S25 | 5.53 | 91.97 | 34.20 | -0.125 |
| GjPAL3 | 77 326.50 | C3428H5470N940O1036S28 | 5.93 | 92.07 | 34.24 | -0.091 |
| GjPAL4 | 77 091.13 | C3406H5453N941O1038S28 | 6.18 | 91.05 | 32.98 | -0.152 |

Fig. 3 Multiple alignment result of PALs from different plant species Characters in the black box represent the enzyme activity center and the red box represent the typical conserved region of the PAL enzyme activity center
| 蛋白 Protein | 长度 Length/aa | 卷曲螺旋结构 Coiled coil structure | 跨膜结构域Transmembrane domain | 磷酸化修饰位点Phosphorylation site | |||||
|---|---|---|---|---|---|---|---|---|---|
| 外→内Outside→Inside | 内→外Inside→Outside | S | T | Y | |||||
| GjPAL1 | 708 | 无 | 3 | 5 | 33 | 21 | 7 | ||
| GjPAL2 | 704 | 有 | 3 | 7 | 34 | 20 | 7 | ||
| GjPAL3 | 711 | 有 | 5 | 6 | 30 | 24 | 8 | ||
| GjPAL4 | 708 | 无 | 2 | 5 | 36 | 19 | 8 | ||
Table 3 Result of transmembrane structure and phospho-rylation sites in GjPALs
| 蛋白 Protein | 长度 Length/aa | 卷曲螺旋结构 Coiled coil structure | 跨膜结构域Transmembrane domain | 磷酸化修饰位点Phosphorylation site | |||||
|---|---|---|---|---|---|---|---|---|---|
| 外→内Outside→Inside | 内→外Inside→Outside | S | T | Y | |||||
| GjPAL1 | 708 | 无 | 3 | 5 | 33 | 21 | 7 | ||
| GjPAL2 | 704 | 有 | 3 | 7 | 34 | 20 | 7 | ||
| GjPAL3 | 711 | 有 | 5 | 6 | 30 | 24 | 8 | ||
| GjPAL4 | 708 | 无 | 2 | 5 | 36 | 19 | 8 | ||

Fig.6 Relative expression of GjPAL genes in different tissues and organs and under different stress treatments A: Different organs. B: SA treatment. C: NaCl treatment. D: PEG treatment. E: Cold stress treatment. F: Phytophthora cryptogea innoculation treatment. “*” indicates significant difference at the 0.05 level, and “**” indicates very significant difference at the 0.01 level
| [1] |
Costa MA, Collins RE, Anterola AM, et al. An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof[J]. Phytochemistry, 2003, 64(6):1097-1112.
doi: 10.1016/S0031-9422(03)00517-X URL |
| [2] |
Huang JL, Gu M, Lai ZB, et al. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress[J]. Plant Physiology, 2010, 153(4):1526-1538.
doi: 10.1104/pp.110.157370 URL |
| [3] |
Ferrer JL, Austin MB, Stewart Jr C, et al. Structure and function of enzymes involved in the biosynjournal of phenylpropanoids[J]. Plant Physiology and Biochemistry, 2008, 46(3):356-370.
doi: 10.1016/j.plaphy.2007.12.009 pmid: 18272377 |
| [4] |
Zhang XB, Liu CJ. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynjournal of phenylpropanoids[J]. Molecular Plant, 2015, 8:17-27.
doi: 10.1016/j.molp.2014.11.001 URL |
| [5] |
Jun SY, Sattler SA, Cortez GS, et al. Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase[J]. Plant Physiology, 2018, 176(2):1452-1468.
doi: 10.1104/pp.17.01608 URL |
| [6] |
De Jong F, Hanley SJ, Beale MH, et al. Characterisation of the willow phenylalanine ammonia-lyase(PAL)gene family reveals expression differences compared with poplar[J]. Phytochemistry, 2015, 117:90-97.
doi: 10.1016/j.phytochem.2015.06.005 URL |
| [7] | 郝向阳, 孙雪丽, 王天池, 等. 植物PAL基因及其编码蛋白的特征与功能研究进展[J]. 热带作物学报, 2018, 39(7):1452-1461. |
| Hao XY, Sun XL, Wang TC, et al. Characteristics and functions of plant Phenylalanine Ammonia Lyase genes and the encoded proteins[J]. Chinese Journal of Tropical Crops, 2018, 39(7):1452-1461. | |
| [8] |
Cochrane FC, Davin LB, Lewis NG. The Arabidopsis phenylalanine ammonia lyase gene family:kinetic characterization of the four PAL isoforms[J]. Phytochemistry, 2004, 65(11):1557-1564.
pmid: 15276452 |
| [9] | 曾嘉丽, 欧阳林娟, 刘家林, 等. 水稻PAL基因的全基因组分析及胁迫表达研究[J]. 基因组学与应用生物学, 2018, 37(9):3881-3888. |
| Ceng JL, Ouyang LJ, Liu JL, et al. Whole genome analysis and stress expression research of PAL gene in rice[J]. Genomics and Applied Biology, 2018, 37(9):3881-3888. | |
| [10] |
Tsai CJ, Harding SA, Tschaplinski TJ, et al. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus[J]. New phytologist, 2006, 172:47-62.
doi: 10.1111/nph.2006.172.issue-1 URL |
| [11] | Gho YS, Kim SJ, Jung KH. Phenylalanine ammonia-lyase family is closely associated with response to phosphate deficiency in rice[J]. Genes & genomics, 2020, 42:67-76. |
| [12] |
Chandrasekaran M, Belachew ST, Yoon E, et al. Expression of β-1, 3-glucanase(GLU)and phenylalanine ammonia-lyase(PAL)genes and their enzymes in tomato plants induced after treatment with Bacillus subtilis CBR05 against Xanthomonas campestris pv. vesicatoria[J]. Journal of General Plant Pathology, 2017, 83:7-13.
doi: 10.1007/s10327-016-0692-5 URL |
| [13] | 熊飞, 卢秦华, 房婉萍, 等. 基于全基因组的茶树PAL家族基因鉴定及其在生物与非生物胁迫下的表达分析[J]. 园艺学报, 2020, 47(3):517-528. |
| Xiong F, Lu QH, Fang WP, et al. Genome-wide Identification and expression analyses of PAL genes under biotic and abiotic stress in Camellia sinensis[J]. Acta Horticulturae Sinica, 2020, 47(3):517-528. | |
| [14] | 杨郁文, 李双, 黄俊宇, 等. 陆地棉苯丙氨酸解氨酶家族基因的鉴定及分析[J]. 分子植物育种, 2017, 15(4):1184-1191. |
| Yang YW, Li S, Huang JY, et al. Identification and analysis of the gene family of phenylalanine ammonialyase in upland cotton[J]. Molecular Plant Breeding, 2017, 15(4):1184-1191. | |
| [15] |
Nugroho LH, Verberne MC, Verpoorte R. Activities of enzymes involved in the phenylpropanoid pathway in constitutively salicylic acid-producing tobacco plants[J]. Plant Physiology and Biochemistry, 2002, 40(9):755-760.
doi: 10.1016/S0981-9428(02)01437-7 URL |
| [16] |
Chaman ME, Copaja SV, Argandoña VH. Relationships between salicylic acid content, phenylalanine ammonia-lyase(PAL)activity, and resistance of barley to aphid infestation[J]. Journal of Agricultural and Food Chemistry, 2003, 51(8):2227-2231.
doi: 10.1021/jf020953b URL |
| [17] |
Ogawa D, Nakajima N, Seo S, et al. The phenylalanine pathway is the main route of salicylic acid biosynjournal in Tobacco mosaic virus-infected tobacco leaves[J]. Plant Biotechnology, 2006, 23(4):395-398.
doi: 10.5511/plantbiotechnology.23.395 URL |
| [18] |
Kim DS, Hwang BK. An important role of the pepper phenylalanine ammonia-lyase gene(PAL1)in salicylic acid-dependent signalling of the defence response to microbial pathogens[J]. Journal of Experimental Botany, 2014, 65(9):2295-2306.
doi: 10.1093/jxb/eru109 URL |
| [19] | 孟祥春, 彭建宗, 王小菁. 光和糖对非洲菊花色素苷积累及CHS、DFR基因表达的影响[J]. 园艺学报, 2007, 34(1):227-230. |
| Meng XC, Peng JZ, Wang XJ. Anthocyanin accumulation and CHS, DFR gene expression regulated by light and sugar in Gerbera hybrid ray floret[J]. Acta Horticulturae Sinica, 2007, 34(1):227-230. | |
| [20] | 王晰, 徐哲, 赖齐贤, 等. 非洲菊切花弯茎影响因素研究进展[J]. 园艺学报, 2015, 42(9):1771-1780. |
| Wang X, Xu Z, Lai QX, et al. Research progress ininfluence factors of stem bending of cut gerbera flower[J]. Acta Horticulturae Sinica, 2015, 42(9):1771-1780. | |
| [21] |
Zhong CM, Tang Y, Pang B, et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida[J]. Horticulture Research, 2020, 7:78.
doi: 10.1038/s41438-020-0296-2 URL |
| [22] | 赖齐贤, 包志毅, 朱祝军, 等. 干旱胁迫对转基因(PSAG12-ipt)非洲菊光合作用的影响[J]. 园艺学报, 2007, 34(1):157-162. |
| Lai QX, Bao ZY, Zhu ZJ, et al. Effects of drought stress on photosynjournal of gerbera modified by PSAG12-ipt[J]. Acta Horticulturae Sinica, 2007, 34(1):157-162. | |
| [23] | 刘芳, 高原, 张竞颐, 等. 非洲菊白粉病病原鉴定及蜡蚧轮枝菌防治试验[J]. 园艺学报, 2010, 37(11):1803-1810. |
| Liu F, Gao Y, Zhang JY, et al. Pathogen identification of gerbera powdery mildew and its control experiment with Verticillium lecanii[J]. Acta Horticulturae Sinica, 2010, 37(11):1803-1810. | |
| [24] | 郝向阳, 林觅, 孙雪丽, 等. 福建非洲菊产区根腐病病原菌的分离与鉴定[J]. 福建农业学报, 2018, 33(4):391-395. |
| Hao XY, Lin M, Sun XL, et al. Isolation and identification of pathogen of gerbera root rot disease in Fujian[J]. Fujian Journal of Agricultural Sciences, 2018, 33(4):391-395. | |
| [25] | Munir N, Cheng CZ, Xia CS, et al. RNA-Seq analysis reveals an essential role of tyrosine metabolism pathway in response to root-rot infection in Gerbera hybrida[J]. PLoS One, 2019, 14(10):e223519. |
| [26] | 郝向阳, 孙雪丽, 刘范, 等. 非洲菊4个POD基因的克隆及表达分析[J]. 西北植物学报, 2018, 38(10):1777-1786. |
| Hao XY, Sun XL, Liu F, et al. Cloning and expression analysis of four gerbera peroxidase genes[J]. Acta Botanica Boreali-Occidentalia Sinica, 2018, 38(10):1777-1786. | |
| [27] |
Ritter H, Schulz GE. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase[J]. The Plant Cell, 2004, 16(12):3426-3436.
doi: 10.1105/tpc.104.025288 URL |
| [28] |
Wang Z, Li JY, Jia CH, et al. Molecular cloning and expression of four phenylalanine ammonia lyase genes from banana interacting with Fusarium oxysporum[J]. Biologia Plantarum, 2016, 60(3):459-468.
doi: 10.1007/s10535-016-0619-1 URL |
| [29] | 许锋, 朱俊, 张风霞, 等. 国槐苯丙氨酸解氨酶基因的克隆、反义表达载体构建及遗传转化[J]. 林业科学研究, 2008, 21(5):611-618. |
| Xu F, Zhu J, Zhang FX, et al. Cloning of PAL gene from Sophora japonica, construction of anti-sense gene of SjPAL and its genetic transformation in Arabidopsis[J]. Forest Research, 2008, 21(5):611-618. | |
| [30] | 许锋, 曹腾, 宁迎晶, 等. 夏枯草苯丙氨酸解氨酶基因的克隆与表达分析[J]. 华北农学报, 2012, 27(1):39-44. |
| Xu F, Cao T, Ning YJ, et al. Molecular cloning and expression analysis of a phenylalanne ammonial-lyase gene from Prunella vulgaris[J]. Acta Agriculturae Boreali-Sinica, 2012, 27(1):39-44. | |
| [31] |
Song J, Wang ZZ. Molecular cloning, expression and characterization of a phenylalanine ammonia-lyase gene(SmPAL1)from Salvia miltiorrhiza[J]. Molecular Biology Reports, 2009, 36(5):939-952.
doi: 10.1007/s11033-008-9266-8 URL |
| [32] |
Shang QM, Li L, Dong CJ. Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L.[J]. Planta, 2012, 236(4):1093-1105.
doi: 10.1007/s00425-012-1659-1 URL |
| [33] | 孙梓健, 汤青林, 宋明, 等. 红叶芥低温胁迫下苯丙氨酸解氨酶活性及其基因的克隆表达[J]. 西南大学学报:自然科学版, 2010, 32(2):90-94. |
| Sun ZJ, Tang QL, Song M, et al. Cloning and expression of PAL gene and PAL activity assay in red-leaf mustard(Brassica juncea var. garrhiza Tsen et Lee)under low temperature stress[J]. Journal of Southwest University:Natural Science Edition, 2010, 32(2):90-94. | |
| [34] |
Morkunas I, Bednarski W, Kopyra M. Defense strategies of pea embryo axes with different levels of sucrose to Fusarium oxysporum and Ascochyta pisi[J]. Physiological and Molecular Plant Pathology, 2008, 72(4):167-178.
doi: 10.1016/j.pmpp.2008.09.003 URL |
| [35] |
Shadle GL, Wesley SV, Korth KL, et al. Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of L-phenylalanine ammonia-lyase[J]. Phytochemistry, 2003, 64:153-161.
pmid: 12946414 |
| [36] |
Chen YP, Li FJ, Tian L, et al. The phenylalanine ammonia lyase gene LjPAL1 is involved in plant defense responses to pathogens and plays diverse roles in Lotus japonicus-rhizobium symbioses[J]. Molecular Plant-Microbe Interactions, 2017, 30(9):739-753.
doi: 10.1094/MPMI-04-17-0080-R URL |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [3] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [4] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [5] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [6] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [7] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [8] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [9] | HOU Rui-ze BAO Yue CHEN Qi-liang MAO Gui-ling WEI Bo-lin HOU Lei-ping LI Mei-lan. Cloning,Expression and Functional Identification of PRR5 Gene in Pakchoi [J]. Biotechnology Bulletin, 2023, 39(10): 128-135. |
| [10] | YANG Min, LONG Yu-qing, ZENG Juan, ZENG Mei, ZHOU Xin-ru, WANG Ling, FU Xue-sen, ZHOU Ri-bao, LIU Xiang-dan. Cloning and Function Analysis of Gene UGTPg17 and UGTPg36 in Lonicera macranthoides [J]. Biotechnology Bulletin, 2023, 39(10): 256-267. |
| [11] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [12] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [13] | GUO Bin-hui, SONG Li. Transcription of Ethylene Biosynthesis and Signaling Associated Genes in Response to Heterodera glycine Infection [J]. Biotechnology Bulletin, 2022, 38(8): 150-158. |
| [14] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| [15] | CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(7): 51-61. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||