








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (11): 238-249.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0167
Previous Articles Next Articles
ZHAO Bao-ding( ), LV Jia, SHEN Yu-yu, GUI Ling, CHEN Zhong-xiu, CHEN Jie, LU Fu-ping, LI Ming(
), LV Jia, SHEN Yu-yu, GUI Ling, CHEN Zhong-xiu, CHEN Jie, LU Fu-ping, LI Ming( )
)
Received:2022-02-15
Online:2022-11-26
Published:2022-12-01
ZHAO Bao-ding, LV Jia, SHEN Yu-yu, GUI Ling, CHEN Zhong-xiu, CHEN Jie, LU Fu-ping, LI Ming. Efficient Transformation of Uridine by Escherichia coli Based on Signal Peptide and Molecular Chaperone Strategy[J]. Biotechnology Bulletin, 2022, 38(11): 238-249.
| 菌株和质粒 Strain and plasmid | 相关特性 Related characteristics | 来源 Source | |
|---|---|---|---|
| 菌株 Strain | E. coli BL21 | Wild type E. coli BL21(DE3) | Lab stock |
| E. coli DH5α | Cloning host | Lab stock | |
| E. coli K12. MG1655 | Wild type | Lab stock | |
| SP-1 | BL21 derivate with plasmid pET22b-Fhud-RihA | This work | |
| SP-2 | BL21 derivate with plasmid pET22b-OmpA-RihA | This work | |
| SP-3 | BL21 derivate with plasmid pET22b-OmpC-RihA | This work | |
| SP-4 | BL21 derivate with plasmid pET22b-OmpF-RihA | This work | |
| SP-5 | BL21 derivate with plasmid pET22b-OmpT-RihA | This work | |
| SP-6 | BL21 derivate with plasmid pET22b-PhoE-RihA | This work | |
| BL21-pCDM4-DnaJ | BL21 derivate with plasmid BL21-pCDM4-DnaJ | This work | |
| BL21-pCDM4-DnaK | BL21 derivate with plasmid BL21-pCDM4-DnaK | This work | |
| BL21-pCDM4-GroEL | BL21 derivate with plasmid BL21-pCDM4-GroEL | This work | |
| BL21-pCDM4-GroES | BL21 derivate with plasmid BL21-pCDM4-GroES | This work | |
| BL21-pCDM4-GrpE | BL21 derivate with plasmid BL21-pCDM4-GrpE | This work | |
| Strain A | BL21 derivate with plasmid pET22b-RihA | This work | |
| Strain B | BL21 derivate with plasmid pET22b-PhoA-RihA | This work | |
| Strain C | BL21 derivate with plasmid pET22b-RihA and pCDM4-GroES-GroEL | This work | |
| Strain D | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-DrpE | This work | |
| Strain E | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-GrpE-GroES-GroEL | This work | |
| Strain F | BL21 derivate with plasmid pET22b-PhoA-RihA and pCDM4-GroES-GroEL | This work | |
| Strain G | BL21 derivate with plasmid pET22b-PhoA-RihA and pCDM4-DnaK-DnaJ-GrpE | This work | |
| Strain H | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-GrpE-GroES-GroEL | This work | |
| 质粒Plasmid | pET22b | T7 promoters,AmpR | Lab stock |
| pCDM4 | T7 promoters,SmR | [ | |
| pET22b-RihA | pET22b derivate with rihA cloned | This work | |
| pET22b-PhoA-RihA | pET22b derivate with PhoA-rihA cloned | This work | |
| pET22b-Fhud-RihA | pET22b derivate with Fhud-rihA cloned | This work | |
| pET22b-OmpA-RihA | pET22b derivate with OmpA-rihA cloned | This work | |
| pET22b-OmpC-RihA | pET22b derivate with OmpC-rihA cloned | This work | |
| pET22b-OmpF-RihA | pET22b derivate with OmpF-rihA cloned | This work | |
| pET22b-OmpT-RihA | pET22b derivate with OmpT-rihA cloned | This work | |
| pET22bpET22b-PhoE-RihA | pET22b derivate with PhoE-rihA cloned | This work | |
| pCDM4-DnaJ | pCDM4 derivate with dnaJ cloned | This work | |
| pCDM4-DnaK | pCDM4 derivate with dnaK cloned | This work | |
| pCDM4-GroEL | pCDM4 derivate with groEL cloned | This work | |
| pCDM4-GroES | pCDM4 derivate with groES cloned | This work | |
| pCDM4-GrpE | pCDM4 derivate with grpE cloned | This work | |
| pCDM4-GroES-GroEL | pCDM4 derivate with groES-groEL cloned | This work | |
| pCDM4-DnaK-DnaJ-GrpE | pCDM4 derivate with dnaK-dnaJ-grpE cloned | This work | |
| pCDM4-DnaK-DnaJ-GrpE- GroES-GroEL | pCDM4 derivate with dnaK-dnaJ-grpE-groES-groEL cloned | This work |
Table 1 Strains and plasmids in this study
| 菌株和质粒 Strain and plasmid | 相关特性 Related characteristics | 来源 Source | |
|---|---|---|---|
| 菌株 Strain | E. coli BL21 | Wild type E. coli BL21(DE3) | Lab stock |
| E. coli DH5α | Cloning host | Lab stock | |
| E. coli K12. MG1655 | Wild type | Lab stock | |
| SP-1 | BL21 derivate with plasmid pET22b-Fhud-RihA | This work | |
| SP-2 | BL21 derivate with plasmid pET22b-OmpA-RihA | This work | |
| SP-3 | BL21 derivate with plasmid pET22b-OmpC-RihA | This work | |
| SP-4 | BL21 derivate with plasmid pET22b-OmpF-RihA | This work | |
| SP-5 | BL21 derivate with plasmid pET22b-OmpT-RihA | This work | |
| SP-6 | BL21 derivate with plasmid pET22b-PhoE-RihA | This work | |
| BL21-pCDM4-DnaJ | BL21 derivate with plasmid BL21-pCDM4-DnaJ | This work | |
| BL21-pCDM4-DnaK | BL21 derivate with plasmid BL21-pCDM4-DnaK | This work | |
| BL21-pCDM4-GroEL | BL21 derivate with plasmid BL21-pCDM4-GroEL | This work | |
| BL21-pCDM4-GroES | BL21 derivate with plasmid BL21-pCDM4-GroES | This work | |
| BL21-pCDM4-GrpE | BL21 derivate with plasmid BL21-pCDM4-GrpE | This work | |
| Strain A | BL21 derivate with plasmid pET22b-RihA | This work | |
| Strain B | BL21 derivate with plasmid pET22b-PhoA-RihA | This work | |
| Strain C | BL21 derivate with plasmid pET22b-RihA and pCDM4-GroES-GroEL | This work | |
| Strain D | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-DrpE | This work | |
| Strain E | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-GrpE-GroES-GroEL | This work | |
| Strain F | BL21 derivate with plasmid pET22b-PhoA-RihA and pCDM4-GroES-GroEL | This work | |
| Strain G | BL21 derivate with plasmid pET22b-PhoA-RihA and pCDM4-DnaK-DnaJ-GrpE | This work | |
| Strain H | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-GrpE-GroES-GroEL | This work | |
| 质粒Plasmid | pET22b | T7 promoters,AmpR | Lab stock |
| pCDM4 | T7 promoters,SmR | [ | |
| pET22b-RihA | pET22b derivate with rihA cloned | This work | |
| pET22b-PhoA-RihA | pET22b derivate with PhoA-rihA cloned | This work | |
| pET22b-Fhud-RihA | pET22b derivate with Fhud-rihA cloned | This work | |
| pET22b-OmpA-RihA | pET22b derivate with OmpA-rihA cloned | This work | |
| pET22b-OmpC-RihA | pET22b derivate with OmpC-rihA cloned | This work | |
| pET22b-OmpF-RihA | pET22b derivate with OmpF-rihA cloned | This work | |
| pET22b-OmpT-RihA | pET22b derivate with OmpT-rihA cloned | This work | |
| pET22bpET22b-PhoE-RihA | pET22b derivate with PhoE-rihA cloned | This work | |
| pCDM4-DnaJ | pCDM4 derivate with dnaJ cloned | This work | |
| pCDM4-DnaK | pCDM4 derivate with dnaK cloned | This work | |
| pCDM4-GroEL | pCDM4 derivate with groEL cloned | This work | |
| pCDM4-GroES | pCDM4 derivate with groES cloned | This work | |
| pCDM4-GrpE | pCDM4 derivate with grpE cloned | This work | |
| pCDM4-GroES-GroEL | pCDM4 derivate with groES-groEL cloned | This work | |
| pCDM4-DnaK-DnaJ-GrpE | pCDM4 derivate with dnaK-dnaJ-grpE cloned | This work | |
| pCDM4-DnaK-DnaJ-GrpE- GroES-GroEL | pCDM4 derivate with dnaK-dnaJ-grpE-groES-groEL cloned | This work |
| 名称 Name | 引物序列Primer sequence(5'-3') |
|---|---|
| pET22bRihAF | cctcgctgcccagccggcgatggccatgGCACTGCCAATTCTGTTAGATTG |
| pET22bRihAR | tgtcgacggagctcgaattcggatccttaAGCGTAAAATTTCAGACGATCAGCC |
| FhuD-RihA F1 | gatgaataccgcccacgcggcggctattgatcccaatatgGCACTGCCAATTCTGTTAGA |
| FhuD F1 | gactgttaacggcgatggcgctttctccgttgttatggcagatgaataccgcccacgcg |
| FhuD F2 | taagaaggagatatacatatgagcggcttacctcttatttcgcgccgtcgactgttaacggcgatggcg |
| OmpA-RihA F1 | gcactggctggtttcgctaccgtagcgcaggccatgGCACTGCCAATTCTGTTAG |
| OmpA F1 | taagaaggagatatacatatgaaaaagacagctatcgcgattgcagtggcactggctggtttcgc |
| OmpC F1 | taagaaggagatatacatatgaaagttaaagtactgtccctcctggtcccagctctgctggtagcag |
| OmpC-RihA F1 | ccagctctgctggtagcaggcgcagcaaacgctatgGCACTGCCAATTCTGTTAGA |
| OmpF F1 | ttaagaaggagatatacatatgatgaagcgcaatattctggcagtgatcgtccctgctctgttagtagcaggt |
| OmpF-RihA F1 | ccctgctctgttagtagcaggtactgcaaacgctatgGCACTGCCAATTCTGTTAG |
| OmpT F1 | tttaagaaggagatatacatatgcgggcgaaacttctgggaatagtcctgacaacccctattgcga |
| OmpT-RihA F1 | cctgacaacccctattgcgatcagctcttttgctatgGCACTGCCAATTCTGTTAG |
| PhoA F1 | tttaagaaggagatatacatatgaaacaaagcactattgcactggcactcttaccgttactgtttacccctgt |
| PhoA-RihA F1 | cttaccgttactgtttacccctgtgacaaaagccatgGCACTGCCAATTCTGTT |
| PhoE F1 | tttaagaaggagatatacatatgaaaaagagcactctggcattagtggtgatgggcattgtggcatct |
| PhoE-RihA F1 | gatgggcattgtggcatctgcatctgtacaggctatgGCACTGCCAATTCTGTTAG |
| dnaJ F1 | GGAATTCCATatggctaagcaagattattacgaga |
| dnaJ R1 | GGACTAGTttagcgggtcaggtcgtca |
| dnaK F1 | GGAATTCCATatgggtaaaataattggtatcgacctgg |
| dnaK R1 | GGACTAGTttattttttgtctttgacttcttcaaattcagc |
| GroEL F1 | GGAATTCCATatggcagctaaagacgtaaaattcg |
| GroEL R1 | GGACTAGTttacatcatgccgcccatgc |
| GroES F1 | GGAATTCCATatgaatattcgtccattgcatgatcg |
| GroES R1 | GGACTAGTttacgcttcaacaattgccagaatgt |
| grpE F1 | GGAATTCCATatgagtagtaaagaacagaaaacgcctg |
| grpE R1 | GGACTAGTttaagcttttgctttcgctacagtaacc |
| Cht F1 | cttgaggggttttttTctagAatcgagatcgatctcgatcccg |
| Cht R1 | actaccggaagcagtgtcgactc |
| To F1 | gatgcgtccggcgtagc |
Table 2 List of primers
| 名称 Name | 引物序列Primer sequence(5'-3') |
|---|---|
| pET22bRihAF | cctcgctgcccagccggcgatggccatgGCACTGCCAATTCTGTTAGATTG |
| pET22bRihAR | tgtcgacggagctcgaattcggatccttaAGCGTAAAATTTCAGACGATCAGCC |
| FhuD-RihA F1 | gatgaataccgcccacgcggcggctattgatcccaatatgGCACTGCCAATTCTGTTAGA |
| FhuD F1 | gactgttaacggcgatggcgctttctccgttgttatggcagatgaataccgcccacgcg |
| FhuD F2 | taagaaggagatatacatatgagcggcttacctcttatttcgcgccgtcgactgttaacggcgatggcg |
| OmpA-RihA F1 | gcactggctggtttcgctaccgtagcgcaggccatgGCACTGCCAATTCTGTTAG |
| OmpA F1 | taagaaggagatatacatatgaaaaagacagctatcgcgattgcagtggcactggctggtttcgc |
| OmpC F1 | taagaaggagatatacatatgaaagttaaagtactgtccctcctggtcccagctctgctggtagcag |
| OmpC-RihA F1 | ccagctctgctggtagcaggcgcagcaaacgctatgGCACTGCCAATTCTGTTAGA |
| OmpF F1 | ttaagaaggagatatacatatgatgaagcgcaatattctggcagtgatcgtccctgctctgttagtagcaggt |
| OmpF-RihA F1 | ccctgctctgttagtagcaggtactgcaaacgctatgGCACTGCCAATTCTGTTAG |
| OmpT F1 | tttaagaaggagatatacatatgcgggcgaaacttctgggaatagtcctgacaacccctattgcga |
| OmpT-RihA F1 | cctgacaacccctattgcgatcagctcttttgctatgGCACTGCCAATTCTGTTAG |
| PhoA F1 | tttaagaaggagatatacatatgaaacaaagcactattgcactggcactcttaccgttactgtttacccctgt |
| PhoA-RihA F1 | cttaccgttactgtttacccctgtgacaaaagccatgGCACTGCCAATTCTGTT |
| PhoE F1 | tttaagaaggagatatacatatgaaaaagagcactctggcattagtggtgatgggcattgtggcatct |
| PhoE-RihA F1 | gatgggcattgtggcatctgcatctgtacaggctatgGCACTGCCAATTCTGTTAG |
| dnaJ F1 | GGAATTCCATatggctaagcaagattattacgaga |
| dnaJ R1 | GGACTAGTttagcgggtcaggtcgtca |
| dnaK F1 | GGAATTCCATatgggtaaaataattggtatcgacctgg |
| dnaK R1 | GGACTAGTttattttttgtctttgacttcttcaaattcagc |
| GroEL F1 | GGAATTCCATatggcagctaaagacgtaaaattcg |
| GroEL R1 | GGACTAGTttacatcatgccgcccatgc |
| GroES F1 | GGAATTCCATatgaatattcgtccattgcatgatcg |
| GroES R1 | GGACTAGTttacgcttcaacaattgccagaatgt |
| grpE F1 | GGAATTCCATatgagtagtaaagaacagaaaacgcctg |
| grpE R1 | GGACTAGTttaagcttttgctttcgctacagtaacc |
| Cht F1 | cttgaggggttttttTctagAatcgagatcgatctcgatcccg |
| Cht R1 | actaccggaagcagtgtcgactc |
| To F1 | gatgcgtccggcgtagc |
| 信号肽 Signal peptide | 来源 Source | 氨基酸序列 Amino acid sequence |
|---|---|---|
| Fhud | Iron(III)hydroxamate ABC transporter periplasmic binding protein | MSGLPLISRRRLLTAMALSPLLWQMNTAHAAAIDPN |
| OmpA | Outer-membrane protein A | MKKTAIAIAVALAGFATVAQA |
| OmpC | Outer-membrane protein C | MKVKVLSLLVPALLVAGAANA |
| OmpF | Outer-membrane protein F | MMKRNILAVIVPALLVAGTANA |
| OmpT | Protease 7 | MRAKLLGIVLTTPIAISSFA |
| PhoA | Alkaline phosphatase | VKQSTIALALLPLLFTPVTKA |
| PhoE | Outer-membrane pore protein F | MKKSTLALVVMGIVASASVQA |
| PelB | Pectin lyase | MKYLLPTAAAGLLLLAAQPAMA |
Table 3 Sequences and sources of signal peptides
| 信号肽 Signal peptide | 来源 Source | 氨基酸序列 Amino acid sequence |
|---|---|---|
| Fhud | Iron(III)hydroxamate ABC transporter periplasmic binding protein | MSGLPLISRRRLLTAMALSPLLWQMNTAHAAAIDPN |
| OmpA | Outer-membrane protein A | MKKTAIAIAVALAGFATVAQA |
| OmpC | Outer-membrane protein C | MKVKVLSLLVPALLVAGAANA |
| OmpF | Outer-membrane protein F | MMKRNILAVIVPALLVAGTANA |
| OmpT | Protease 7 | MRAKLLGIVLTTPIAISSFA |
| PhoA | Alkaline phosphatase | VKQSTIALALLPLLFTPVTKA |
| PhoE | Outer-membrane pore protein F | MKKSTLALVVMGIVASASVQA |
| PelB | Pectin lyase | MKYLLPTAAAGLLLLAAQPAMA |
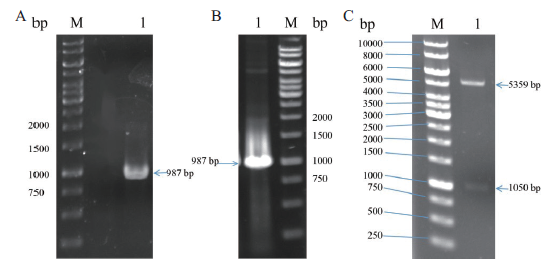
Fig. 3 Construction and validation of pET22b-RihA plasmid A:PCR amplification product of rihA(M:DNA marker,1:rihA). B:Colony PCR validation of recombinant plasmid pET22b-RihA(M:DNA marker,1:rih A). C:Double digestion verification of recombinant plasmid pET22b-RihA(M:DNA marker,1:pET22b-RihA)
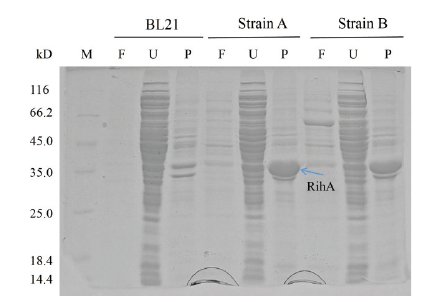
Fig.4 SDS-PAGE analysis of expression products of strain A,B and BL21 M:Molecular weight marker;F:fermentation supernatant;U:ultrasonication supernatant;P:precipitation
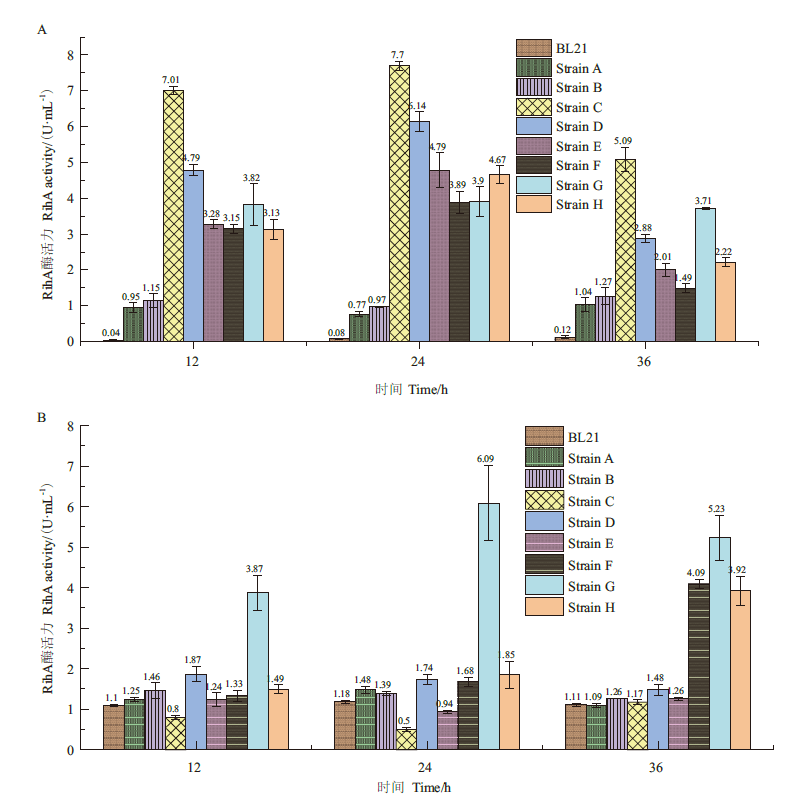
Fig.8 Extracellular and intracellular RihA activities of 9 strains at different time A:Extracellular RihA enzyme activity. B:Intracellular RihA enzyme activity
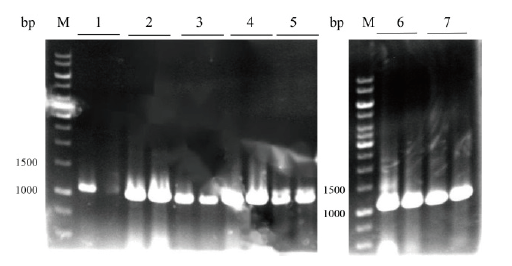
Fig.6 Colony PCR verification of seven kinds of signal pep-tide plasmid pET22b-Fhud/OmpA/OmpC/OmpF/OmpT/PhoA/PhoE-RihA M:DNA marker;1:Fhud-rihA(1 088 bp);2:OmpA-rihA(1 043 bp);3:OmpC-rihA(1 043 bp);4:OmpF-rihA(1 047 bp);5:OmpT-rihA(1 042 bp);6:PhoA-rihA(1 045 bp);7:PhoE-rihA(1 045 bp)
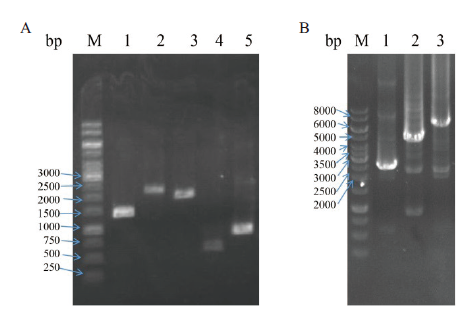
Fig.8 Construction and validation of tandem molecular chaperone plasmids A:PCR amplification of different chaperone expression boxes,M:DNA marker,1:T7P-dnaJ-T7T(1 401 bp),2:T7P-dnaK-T7T(2 187 bp),3:T7P-groEL-T7T(1 917 bp),4:T7P-groES-T7T(564 bp),5:T7P-grpE-T7T(864 bp). B:PCR validation of tandem molecular chaperone plasmids,M:DNA marker,1:groES-groEL(2 429 bp),2:dnaK-dnaJ-grpE(4 348 bp),3:dnaK-dnaJ-grpE-groES-groEL(6 725 bp)

Fig.9 SDS-PAGE analysis of expression products of differ-ent strains M:Molecular weight marker;F:fermentation supernatant;U:ultrasonication supernatant;P:precipitation
| 投底物量 Amount of the substrate | 发酵液中初始尿苷浓度 Initial uridine concentration of fermentation broth/(g·L-1) | 尿嘧啶产量 Uracil yield/(g·L-1) | 尿嘧啶产率 Uracil productive rate /% |
|---|---|---|---|
| 1倍体积/20 mL One time the volume/20 mL | 163.02±4.50 | 73.45±1.03 | 98.16±1.38 |
| 1.5倍体积/30 mL 1.5 times the volume/30 mL | 195.60±6.01 | 88.09±1.33 | 98.12±1.48 |
| 2倍体积/40 mL Two times the volume/40 mL | 218.78±3.95 | 97.90±2.17 | 97.49±2.16 |
Table 4 Uracil yield and uracil productive rate transformed with different substrate concentration
| 投底物量 Amount of the substrate | 发酵液中初始尿苷浓度 Initial uridine concentration of fermentation broth/(g·L-1) | 尿嘧啶产量 Uracil yield/(g·L-1) | 尿嘧啶产率 Uracil productive rate /% |
|---|---|---|---|
| 1倍体积/20 mL One time the volume/20 mL | 163.02±4.50 | 73.45±1.03 | 98.16±1.38 |
| 1.5倍体积/30 mL 1.5 times the volume/30 mL | 195.60±6.01 | 88.09±1.33 | 98.12±1.48 |
| 2倍体积/40 mL Two times the volume/40 mL | 218.78±3.95 | 97.90±2.17 | 97.49±2.16 |
| [1] |
Ganyecz Á, Kállay M, Csontos J. Thermochemistry of uracil, thymine, cytosine, and adenine[J]. J Phys Chem A, 2019, 123(18):4057-4067.
doi: 10.1021/acs.jpca.9b02061 pmid: 30977653 |
| [2] | Ramesh D, Vijayakumar BG, Kannan T. Therapeutic potential of uracil and its derivatives in countering pathogenic and physiological disorders[J]. Eur J Med Chem, 2020, 207:112801. |
| [3] |
Pałasz A, Cież D. In search of uracil derivatives as bioactive agents. Uracils and fused uracils:Synthesis, biological activity and applications[J]. Eur J Med Chem, 2015, 97:582-611.
doi: 10.1016/j.ejmech.2014.10.008 URL |
| [4] | 王利敏, 王思瑶, 张诗缇, 等. 医药中间体尿嘧啶的合成研究[J]. 辽宁医学院学报, 2014, 35(6):7-9. |
| Wang LM, Wang SY, Zhang ST, et al. Study on the synthesis of uracil as a pharmaceutical intermediate[J]. J Liaoning Med Univ, 2014, 35(6):7-9. | |
| [5] |
Kim S, Lee WJ, Song I, et al. Production of uracil from methane by a newly isolated Methylomonas sp. SW1[J]. J Biotechnol, 2016, 240:43-47.
doi: 10.1016/j.jbiotec.2016.10.019 URL |
| [6] |
Lee WJ, Kim S, Song I, et al. Microbial production of uracil by an isolated Methylobacterium sp. WJ4 using methanol[J]. Enzyme Microb Technol, 2018, 111:63-66.
doi: 10.1016/j.enzmictec.2017.10.003 URL |
| [7] |
Petersen C, Møller LB. The RihA RihB, and RihC ribonucleoside hydrolases of Escherichia coli. substrate specificity, gene expression, and regulation[J]. J Biol Chem, 2001, 276(2):884-894.
doi: 10.1074/jbc.M008300200 pmid: 11027694 |
| [8] |
Versées W, Steyaert J. Catalysis by nucleoside hydrolases[J]. Curr Opin Struct Biol, 2003, 13(6):731-738.
doi: 10.1016/j.sbi.2003.10.002 URL |
| [9] |
Yoon SH, Kim SK, Kim JF. Secretory production of recombinant proteins in Escherichia coli[J]. Recent Pat Biotechnol, 2010, 4(1):23-29.
doi: 10.2174/187220810790069550 URL |
| [10] | Mergulhão FJ, Monteiro GA. Periplasmic targeting of recombinant proteins in Escherichia coli[M]// van der Giezen M. Protein Targeting Protocols. Totowa, NJ: Humana Press, 2007:47-61. |
| [11] | Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli:advances and challenges[J]. Front Microbiol, 2014, 5:172. |
| [12] |
Singhvi P, Saneja A, Srichandan S, et al. Bacterial inclusion bodies:a treasure trove of bioactive proteins[J]. Trends Biotechnol, 2020, 38(5):474-486.
doi: 10.1016/j.tibtech.2019.12.011 URL |
| [13] |
Kleiner-Grote GRM, Risse JM, Friehs K. Secretion of recombinant proteins from E. coli[J]. Eng Life Sci, 2018, 18(8):532-550.
doi: 10.1002/elsc.201700200 pmid: 32624934 |
| [14] |
Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli[J]. Nat Biotechnol, 2004, 22(11):1399-1408.
doi: 10.1038/nbt1029 URL |
| [15] |
Oganesyan N, Ankoudinova I, Kim SH, et al. Effect of osmotic stress and heat shock in recombinant protein overexpression and crystallization[J]. Protein Expr Purif, 2007, 52(2):280-285.
doi: 10.1016/j.pep.2006.09.015 URL |
| [16] |
Fatima K, Naqvi F, Younas H. A review:molecular chaperone-mediated folding, unfolding and disaggregation of expressed recombinant proteins[J]. Cell Biochem Biophys, 2021, 79(2):153-174.
doi: 10.1007/s12013-021-00970-5 pmid: 33634426 |
| [17] |
Tian YX, Chen J, Yu HM, et al. Overproduction of the Escherichia coli chaperones GroEL-GroES in Rhodococcus ruber improves the activity and stability of cell catalysts harboring a nitrile hydratase[J]. J Microbiol Biotechnol, 2016, 26(2):337-346.
doi: 10.4014/jmb.1509.09084 URL |
| [18] | Yao D, Fan J, Han RZ, et al. Enhancing soluble expression of sucrose phosphorylase in Escherichia coli by molecular chaperones[J]. Protein Expr Purif, 2020, 169:105571. |
| [19] |
Pan D, Zha X, Yu XH, et al. Enhanced expression of soluble human papillomavirus L1 through coexpression of molecular chaperonin in Escherichia coli[J]. Protein Expr Purif, 2016, 120:92-98.
doi: 10.1016/j.pep.2015.12.016 URL |
| [20] | 邓通, 周海胜, 吴坚平, 等. 基于分子伴侣策略提高NADPH依赖型醇脱氢酶的异源可溶性表达[J]. 中国生物工程杂志, 2020, 40(8):24-32. |
| Deng T, Zhou HS, Wu JP, et al. Enhance soluble heteroexpression of a NADPH-dependent alcohol dehydrogenase based on the chaperone strategy[J]. China Biotechnol, 2020, 40(8):24-32. | |
| [21] |
Low KO, Muhammad Mahadi N, Md Illias R. Optimisation of signal peptide for recombinant protein secretion in bacterial hosts[J]. Appl Microbiol Biotechnol, 2013, 97(9):3811-3826.
doi: 10.1007/s00253-013-4831-z pmid: 23529680 |
| [22] |
Mirzadeh K, Shilling PJ, Elfageih R, et al. Increased production of periplasmic proteins in Escherichia coli by directed evolution of the translation initiation region[J]. Microb Cell Fact, 2020, 19(1):85.
doi: 10.1186/s12934-020-01339-8 pmid: 32264894 |
| [23] |
Brockmeier U, Caspers M, Freudl R, et al. Systematic screening of all signal peptides from Bacillus subtilis:a powerful strategy in optimizing heterologous protein secretion in Gram-positive bacteria[J]. J Mol Biol, 2006, 362(3):393-402.
pmid: 16930615 |
| [24] |
Mathiesen G, Sveen A, Brurberg MB, et al. Genome-wide analysis of signal peptide functionality in Lactobacillus plantarum WCFS1[J]. BMC Genomics, 2009, 10:425.
doi: 10.1186/1471-2164-10-425 pmid: 19744343 |
| [25] |
Freudl R. Signal peptides for recombinant protein secretion in bacterial expression systems[J]. Microb Cell Fact, 2018, 17(1):52.
doi: 10.1186/s12934-018-0901-3 pmid: 29598818 |
| [26] |
Xu P, Vansiri A, Bhan N, et al. ePathBrick:a synthetic biology platform for engineering metabolic pathways in E. coli[J]. ACS Synth Biol, 2012, 1(7):256-266.
doi: 10.1021/sb300016b URL |
| [27] |
Freilich R, Arhar T, Abrams JL, et al. Protein-protein interactions in the molecular chaperone network[J]. Acc Chem Res, 2018, 51(4):940-949.
doi: 10.1021/acs.accounts.8b00036 URL |
| [28] |
Tang YC, Chang HC, Roeben A, et al. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein[J]. Cell, 2006, 125(5):903-914.
doi: 10.1016/j.cell.2006.04.027 URL |
| [29] |
Clare DK, Bakkes PJ, van Heerikhuizen H, et al. Chaperonin complex with a newly folded protein encapsulated in the folding chamber[J]. Nature, 2009, 457(7225):107-110.
doi: 10.1038/nature07479 URL |
| [30] |
Mayer MP, Bukau B. Hsp70 chaperones:cellular functions and molecular mechanism[J]. Cell Mol Life Sci, 2005, 62(6):670-684.
pmid: 15770419 |
| [31] |
Deuerling E, Schulze-Specking A, Tomoyasu T, et al. Trigger factor and DnaK cooperate in folding of newly synthesized proteins[J]. Nature, 1999, 400(6745):693-696.
doi: 10.1038/23301 URL |
| [32] |
Genevaux P, Keppel F, Schwager F, et al. In vivo analysis of the overlapping functions of DnaK and trigger factor[J]. EMBO Rep, 2004, 5(2):195-200.
pmid: 14726952 |
| [33] |
Garavito MF, Narváez-Ortiz HY, Zimmermann BH. Pyrimidine metabolism:dynamic and versatile pathways in pathogens and cellular development[J]. Journal of genetics and genomics, 2015, 42(5):195-205.
doi: 10.1016/j.jgg.2015.04.004 pmid: 26059768 |
|
Garavito MF, Narváez-Ortiz HY, Zimmermann BH. Pyrimidine metabolism:dynamic and versatile pathways in pathogens and cellular development[J]. J Genet Genomics, 2015, 42(5):195-205.
doi: 10.1016/j.jgg.2015.04.004 pmid: 26059768 |
|
| [34] |
West TP. Isolation and characterization of an Escherichia coli B mutant strain defective in uracil catabolism[J]. Can J Microbiol, 1998, 44(11):1106-1109.
pmid: 10030006 |
| [1] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [2] | DONG Cong, GAO Qing-hua, WANG Yue, LUO Tong-yang, WANG Qing-qing. Increasing the Expression of FAD-dependent Glucose Dehydrogenase by Recombinant Pichia pastoris Using a Combined Strategy [J]. Biotechnology Bulletin, 2023, 39(6): 316-324. |
| [3] | DUAN Xu-guo, ZHANG Yu-hua, HUANG Ting-ting, DING Qian, LUAN Shu-yue, ZHU Qiu-yu. Synergetic Enhancing the Soluble Expression of Thermotoga maritima α-Glucan Phosphorylase by Chemical Chaperones and Induction Condition Optimization [J]. Biotechnology Bulletin, 2021, 37(8): 233-242. |
| [4] | MIAO Hua-biao, CAO Yan, YANG Meng-han, HUANG Zun-xi. The Strategy for Enhancing Foreign Proteins Expression by Signal Peptide in Bacillus subtilis [J]. Biotechnology Bulletin, 2021, 37(6): 259-271. |
| [5] | MIN Qi, GAO Zi-han, YAO Yin, ZHANG Hua-shan, XIONG Hai-rong, ZHANG Li. Effect of Co-expression of HAC1 and Molecular Chaperone Genes on the Expression of Mannanase in Pichia pastoris [J]. Biotechnology Bulletin, 2020, 36(5): 159-168. |
| [6] | LIANG Xin-xin, TANG Dan, HUO Yi-xin. Green Biotransformation of Protein-derived Biomass [J]. Biotechnology Bulletin, 2020, 36(12): 216-228. |
| [7] | ZHANG Yu-wen, YUAN Hang, YU Jiang-yue, MA Xiao-xiao, SHI Chao-shuo, LI Yu. Screening of a Bacterial Strain Efficiently Degrading Feather Waste and Optimization of Its Expression Condition [J]. Biotechnology Bulletin, 2019, 35(9): 93-98. |
| [8] | GUO Lei-zhou, HAN Jia-hui, TANG Yin, LI Jiang, HUANG Cheng, DAI Qi-lin, WANG Jin, PING Shu-zhen, JIANG Shi-jie. Effect of Signal Peptide-like Sequence in DrwH on Its Antioxidant Function [J]. Biotechnology Bulletin, 2019, 35(5): 125-132. |
| [9] | ZHAO Xiang-jie, YANG Wen-jun, YANG Rong-ling, WU Ting-ting, WANG Zhao-yu, XU Ning-ning, HE Jia-mei. Research Progress on Biotransformation Modification of Anthocyanins [J]. Biotechnology Bulletin, 2019, 35(10): 205-211. |
| [10] | YANG He-bao ,HU Mei-rong ,ZHENG Xiang ,MOU Qing-xuan ,GAO Pei-ru. Effects of Different Signal Peptides and Their Molecular Chaperones on the Secretion of Neutral Protease in Bacillus subtilis [J]. Biotechnology Bulletin, 2018, 34(6): 134-140. |
| [11] | ZHANG Heng, LIU Ying-ying, CHEN Yun, PING Shu-zhen, WANG Jin. Biological Identification of Dgl5 in Deinococcus gobiensis I-0 [J]. Biotechnology Bulletin, 2018, 34(3): 177-184. |
| [12] | WANG Zhi-wen, CHEN Hai-bo, SONG Fu-ping, GUO Shu-yuan. Identification and Analysis of Secretory Proteins in Bacillus thuringiensis [J]. Biotechnology Bulletin, 2017, 33(4): 169-176. |
| [13] | ZHU Feng-zhi, CHENG Cheng, LIU Xiang-sheng, ZHANG Kun WANG, Li-shuang ,WANG Min, LUO Jian-mei. Optimization of Biotransformation Technology for 10-DAB Production from Baccatin III Using Response Surface Methodology [J]. Biotechnology Bulletin, 2017, 33(4): 238-246. |
| [14] | HE Qing-lin, BAI Yan-fen, ZHOU Wei, YIN Hua, CHEN Ning, ZHUANG Yi-bin, LIU Tao. Biocatalysis of Phenolic Glycosides Natural Products in Escherichia coli Strain Using UGT73B6 [J]. Biotechnology Bulletin, 2017, 33(11): 136-142. |
| [15] | CAI Dong-mei, GONG Guo-li. The Current Status and Future Perspectives of Production of Biopharmaceuticals in Escherichia coli [J]. Biotechnology Bulletin, 2016, 32(8): 34-40. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||