








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (11): 269-276.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0329
Previous Articles Next Articles
ZHANG Chen1,2( ), ZHANG Tong-tong2,3, LIU Hai-ping2,3(
), ZHANG Tong-tong2,3, LIU Hai-ping2,3( )
)
Received:2022-03-19
Online:2022-11-26
Published:2022-12-01
Contact:
LIU Hai-ping
E-mail:zhangchen@tib.cas.cn;liu_hp@tib.cas.cn
ZHANG Chen, ZHANG Tong-tong, LIU Hai-ping. Screening and Identification of Ethylene-forming Enzymes with High Activity and Thermostability[J]. Biotechnology Bulletin, 2022, 38(11): 269-276.
| 蛋白名称 Protein | 种属名称 Species | 蛋白分子量 Molecular weight/kD |
|---|---|---|
| CgEFE | Colletotrichum gloeosporioides | 46.94 |
| ChEFE | Colletotrichum higginsianum | 47.00 |
| GgEFE | Glomerella graminicola | 46.30 |
| SbEFF | Streptomyces bottropensis | 38.52 |
| StEFE | Streptomyces turgidiscabies | 38.55 |
| SsvEFE | Streptomyces sviceus | 39.61 |
Table 1 Species of EFEs
| 蛋白名称 Protein | 种属名称 Species | 蛋白分子量 Molecular weight/kD |
|---|---|---|
| CgEFE | Colletotrichum gloeosporioides | 46.94 |
| ChEFE | Colletotrichum higginsianum | 47.00 |
| GgEFE | Glomerella graminicola | 46.30 |
| SbEFF | Streptomyces bottropensis | 38.52 |
| StEFE | Streptomyces turgidiscabies | 38.55 |
| SsvEFE | Streptomyces sviceus | 39.61 |
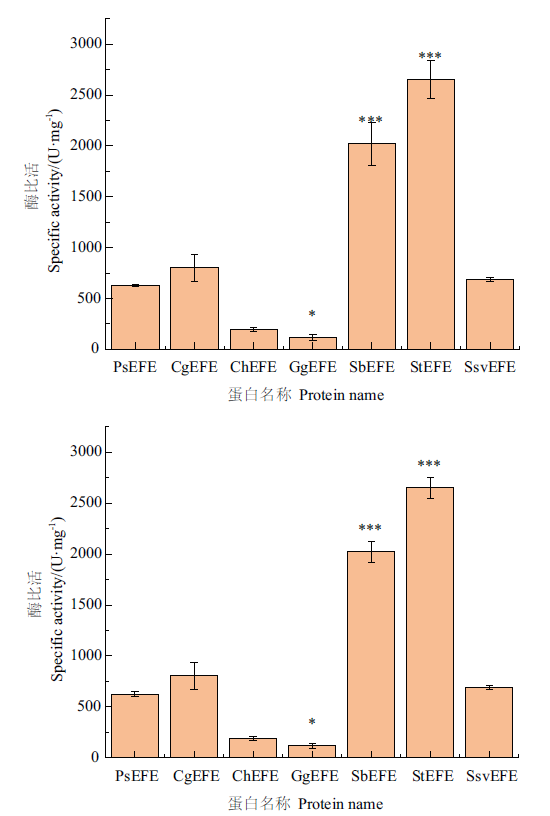
Fig. 2 Comparison of the specific activities of EFEs from various species The error bars represent the standard deviation,* represents significant difference(P<0.05),and *** represents extremely significant difference(P<0.001),The same below
| 蛋白Protein | 底物 Substrate | Km/(μmol·L-1) | Kcat/min-1 | Kcat/Km /(L·μmmol-1·min-1) |
|---|---|---|---|---|
| PsEFE | 2-OG1 | 26.41±10.06 | 39.55±5.86 | 1.50 |
| L-Arg1 | 38.18±4.79 | 35.23±1.86 | 0.92 | |
| L-Arg2 | 101.30±31.60 | 0.92±0.14 | 0.009 | |
| SbEFE | 2-OG1 | 116.60±23.66 | 145.20±15.33 | 1.25 |
| L-Arg1 | 27.26±5.17 | 135.35±9.29 | 4.97 | |
| L-Arg2 | 96.16±32.68 | 1.70±0.27 | 0.018 | |
| StEFE | 2-OG1 | 261.30±192.40 | 298.78±149.68 | 1.14 |
| L-Arg1 | 24.04±8.29 | 132.82±17.54 | 5.52 | |
| L-Arg2 | 92.89±34.28 | 1.61±0.28 | 0.017 |
Table 2 Kinetic parameters of SbEFE and StEFE for subs-trate 2-OG and L-Arg
| 蛋白Protein | 底物 Substrate | Km/(μmol·L-1) | Kcat/min-1 | Kcat/Km /(L·μmmol-1·min-1) |
|---|---|---|---|---|
| PsEFE | 2-OG1 | 26.41±10.06 | 39.55±5.86 | 1.50 |
| L-Arg1 | 38.18±4.79 | 35.23±1.86 | 0.92 | |
| L-Arg2 | 101.30±31.60 | 0.92±0.14 | 0.009 | |
| SbEFE | 2-OG1 | 116.60±23.66 | 145.20±15.33 | 1.25 |
| L-Arg1 | 27.26±5.17 | 135.35±9.29 | 4.97 | |
| L-Arg2 | 96.16±32.68 | 1.70±0.27 | 0.018 | |
| StEFE | 2-OG1 | 261.30±192.40 | 298.78±149.68 | 1.14 |
| L-Arg1 | 24.04±8.29 | 132.82±17.54 | 5.52 | |
| L-Arg2 | 92.89±34.28 | 1.61±0.28 | 0.017 |
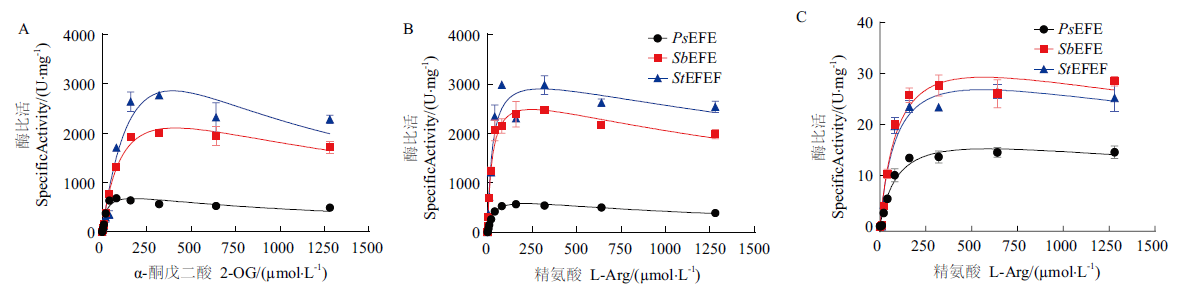
Fig. 7 Enzyme activities of PsEFE,SbEFE and StEFE A:Ethylene production by PsEFE,SbEFE and StEFE with different 2-OG concentration. B:Ethylene production by PsEFE,SbEFE and StEFE with different L-Arg concentration. C:P5C activity curve of PsEFE,SbEFE and StEFE with L-Arg concentration
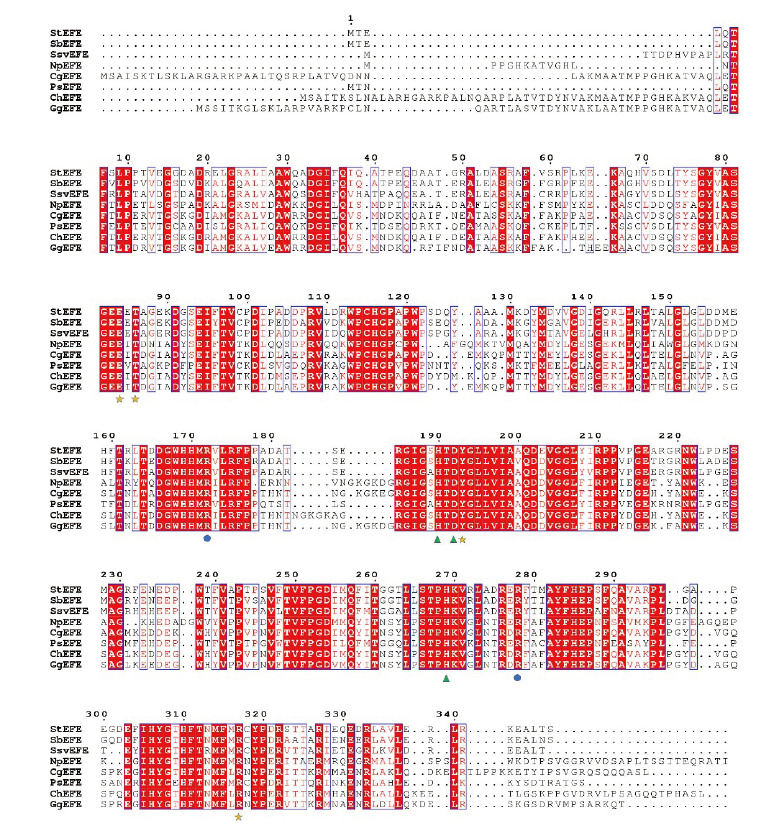
Fig. 8 Sequence alignment of EFEs ▲represents amino acids involved in metal chelation. ●represents amino acids involved in 2-OG binding,★represents amino acids involved in L-Arg binding
| [1] | 夏春晖, 裴仁彦, 王辉, 等. 乙烯技术发展现状及展望[J]. 煤炭与化工, 2020, 43(1):131-134, 138. |
| Xia CH, Pei RY, Wang H, et al. Development status and prospects of ethylene technology[J]. Coal Chem Ind, 2020, 43(1):131-134, 138. | |
| [2] | Fan D, Dai DJ, Wu HS. Ethylene formation by catalytic dehydration of ethanol with industrial considerations[J]. Materials(Basel), 2012, 6(1):101-115. |
| [3] |
Biale JB. Effect of emanations from several species of fungi on respiration and color development of Citrus fruits[J]. Science, 1940, 91(2367):458-459.
pmid: 17838924 |
| [4] |
Schaller GE. Ethylene action in plants[J]. Ann Bot, 2007, 99(3):561.
doi: 10.1093/aob/mcm004 URL |
| [5] | Wang KLC, Li H, Ecker JR. Ethylene biosynthesis and signaling networks[J]. Plant Cell, 2002, 14(Suppl):S131-S151. |
| [6] |
Ghanta M, Fahey D, Subramaniam B. Environmental impacts of ethylene production from diverse feedstocks and energy sources[J]. Appl Petrochem Res, 2014, 4(2):167-179.
doi: 10.1007/s13203-013-0029-7 URL |
| [7] | Dickey DS. Technology profile:‘green’ ethylene production[J]. Chemical Engineering, 2015, 122:40-47. |
| [8] |
Eckert C, Xu W, Xiong W, et al. Ethylene-forming enzyme and bioethylene production[J]. Biotechnol Biofuels, 2014, 7(1):33.
doi: 10.1186/1754-6834-7-33 pmid: 24589138 |
| [9] |
Fukuda H, Ogawa T, Ishihara K, et al. Molecular cloning in Escherichia coli, expression, and nucleotide sequence of the gene for the ethylene-forming enzyme of Pseudomonas syringae pv. phaseolicola PK2[J]. Biochem Biophys Res Commun, 1992, 188(2):826-832.
doi: 10.1016/0006-291X(92)91131-9 URL |
| [10] |
Ishihara K, Matsuoka M, Inoue Y, et al. Overexpression and in vitro reconstitution of the ethylene-forming enzyme from Pseudomonas syringae[J]. J Ferment Bioeng, 1995, 79(3):205-211.
doi: 10.1016/0922-338X(95)90604-X URL |
| [11] |
Young RE, Pratt HK, Biale JB. Identification of ethylene as a volatile product of the fungus Penicillium digitatum[J]. Plant Physiol, 1951, 26(2):304-310.
doi: 10.1104/pp.26.2.304 pmid: 16654368 |
| [12] |
Lynch S, Eckert C, Yu JP, et al. Overcoming substrate limitations for improved production of ethylene in E. coli[J]. Biotechnol Biofuels, 2016, 9:3.
doi: 10.1186/s13068-015-0413-x pmid: 26734073 |
| [13] |
Pirkov I, Albers E, Norbeck J, et al. Ethylene production by metabolic engineering of the yeast Saccharomyces cerevisiae[J]. Metab Eng, 2008, 10(5):276-280.
doi: 10.1016/j.ymben.2008.06.006 pmid: 18640286 |
| [14] | Chen X, Liang Y, Hua J, et al. Overexpression of bacterial ethylene-forming enzyme gene in Trichoderma reesei enhanced the production of ethylene[J]. Int J Biol Sci, 2010:96-106. |
| [15] |
Fukuda H, Sakai M, Nagahama K, et al. Heterologous expression of the gene for the ethylene-forming enzyme from Pseudomonas syringae in the cyanobacteriumSynechococcus[J]. Biotechnol Lett, 1994, 16(1):1-6.
doi: 10.1007/BF01022614 URL |
| [16] |
Ungerer J, Tao L, Davis M, et al. Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803[J]. Energy Environ Sci, 2012, 5(10):8998.
doi: 10.1039/c2ee22555g URL |
| [17] |
Zhu T, Xie XM, Li ZM, et al. Enhancing photosynthetic production of ethylene in genetically engineered Synechocystis sp. PCC 6803[J]. Green Chem, 2015, 17(1):421-434.
doi: 10.1039/C4GC01730G URL |
| [18] |
Veetil VP, Angermayr SA, Hellingwerf KJ. Ethylene production with engineered Synechocystis sp PCC 6803 strains[J]. Microb Cell Fact, 2017, 16(1):34.
doi: 10.1186/s12934-017-0645-5 URL |
| [19] |
Carbonell V, Vuorio E, Aro EM, et al. Enhanced stable production of ethylene in photosynthetic cyanobacterium Synechococcus elongatus PCC 7942[J]. World J Microbiol Biotechnol, 2019, 35(5):77.
doi: 10.1007/s11274-019-2652-7 URL |
| [20] | Guerrero F, Carbonell V, Cossu M, et al. Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC 6803[J]. PLoS One, 2012, 7(11):e50470. |
| [21] |
Wang B, Eckert C, Maness PC, et al. A genetic toolbox for modulating the expression of heterologous genes in the cyanobacterium Synechocystis sp. PCC 6803[J]. ACS Synth Biol, 2018, 7(1):276-286.
doi: 10.1021/acssynbio.7b00297 pmid: 29232504 |
| [22] |
Nagahama K, Ogawa T, Fujii T, et al. Purification and properties of an ethylene-forming enzyme from Pseudomonas syringae pv. phaseolicola PK2[J]. J Gen Microbiol, 1991, 137(10):2281-2286.
pmid: 1770346 |
| [23] |
Nagahama K, Yoshino K, Matsuoka M, et al. Site-directed mutagenesis of histidine residues in the ethylene-forming enzyme from Pseudomonas syringae[J]. J Ferment Bioeng, 1998, 85(3):255-258.
doi: 10.1016/S0922-338X(97)85671-1 URL |
| [24] | 苏宾宾, 张佟佟, 刘海萍. 一种高活性乙烯合成酶的鉴定和性质研究[J]. 生物学杂志, 2021, 38(4):54-58. |
| Su BB, Zhang TT, Liu HP. Identification and characterization of a highly active ethylene forming enzyme[J]. J Biol, 2021, 38(4):54-58. | |
| [25] |
Martinez S, Hausinger RP. Biochemical and spectroscopic characterization of the non-heme Fe(II)- and 2-oxoglutarate-dependent ethylene-forming enzyme from Pseudomonas syringae pv. phaseolicola PK2[J]. Biochemistry, 2016, 55(43):5989-5999.
pmid: 27749027 |
| [26] |
Petraco NDK, Proni G, Jackiw JJ, et al. Amino acid alanine reactivity with the fingerprint reagent ninhydrin. A detailed ab initio computational study[J]. J Forensic Sci, 2006, 51(6):1267-1275.
doi: 10.1111/j.1556-4029.2006.00271.x URL |
| [27] |
Ravikumar H, Devaraju KS, Shetty KT. Effect of pH on spectral characteristics of P5C-ninhydrin derivative:application in the assay of ornithine amino transferase activity from tissue lysate[J]. Indian J Clin Biochem, 2008, 23(2):117-122.
doi: 10.1007/s12291-008-0028-0 URL |
| [28] |
Zhang ZH, Smart TJ, Choi H, et al. Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate[J]. PNAS, 2017, 114(18):4667-4672.
doi: 10.1073/pnas.1617760114 pmid: 28420789 |
| [29] |
Martinez S, Fellner M, Herr CQ, et al. Structures and mechanisms of the non-heme Fe(II)- and 2-oxoglutarate-dependent ethylene-forming enzyme:substrate binding creates a twist[J]. J Am Chem Soc, 2017, 139(34):11980-11988.
doi: 10.1021/jacs.7b06186 pmid: 28780854 |
| [30] |
Notredame C, Higgins DG, Heringa J. T-Coffee:a novel method for fast and accurate multiple sequence alignment[J]. J Mol Biol, 2000, 302(1):205-217.
doi: 10.1006/jmbi.2000.4042 pmid: 10964570 |
| [31] | Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server[J]. Nucleic Acids Res, 2014, 42(Web Server issue):W320-W324. |
| [1] | PAN Guo-qiang, WU Si-yuan, LIU Lu, GUO Hui-ming, CHENG Hong-mei, SU Xiao-feng. Construction and Preliminary Analysis of Verticillim dahliae Mutant Library [J]. Biotechnology Bulletin, 2023, 39(5): 112-119. |
| [2] | YANG Jun-zhao, ZHANG Xin-rui, SUN Qing-yang, ZHENG Fei. Affecting Mechanism of Loop B3 on the Function of GH7 Endoglucanase [J]. Biotechnology Bulletin, 2023, 39(10): 281-291. |
| [3] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [4] | ZHANG Xue, TAN Yu-meng, JIANG Hai-xia, YANG Guang-yu. Directed Evolution of α-1,2-fucosyltransferase by a Single-cell Ultra-high-throughput Screening Method [J]. Biotechnology Bulletin, 2022, 38(1): 289-298. |
| [5] | WANG Lu-lu, GENG Xing-min, XU Shi-da. Ethylene Receptor in Fruit Ripening and Flower Senescence [J]. Biotechnology Bulletin, 2021, 37(3): 144-152. |
| [6] | CHEN Chun, SU Ling-qia, XIA Wei, WU Jing. Improved the Thermostability of MTHase from Arthrobacter ramosus by Directed Evolution [J]. Biotechnology Bulletin, 2021, 37(3): 84-91. |
| [7] | WANG Qi-yuan, WANG Jia-chen, YE Lei, JIANG Fan. Research Advances on Enhancement of Plant Resistance to Salinity Stress by Rhizobacteria Containing ACC Deaminase [J]. Biotechnology Bulletin, 2021, 37(2): 174-186. |
| [8] | WU Jiao, YU Gui-zhen, YUAN Hang, LIU Xian, GAO Yan-xiu, GONG Ming, ZOU Zhu-rong. Improvement on the Thermostability of Target Proteins by Fusing Rubredoxin from Hyperthermophile Pyrococcus furiosus [J]. Biotechnology Bulletin, 2021, 37(10): 110-119. |
| [9] | GAO Chao, HAO Kong-li, ZHAO Yu-ting, MAO Ying-xiang, CHI Ming-yan, ZHANG Jie. Identification and Analysis of a Strain of Enterobacter hormaechei Capable of Degrading Polyethylene [J]. Biotechnology Bulletin, 2020, 36(10): 99-104. |
| [10] | SHI Li-xia, GAO Song-feng, ZHU Lei-lei. Research Advance in Polyethylene Terephthalate Hydrolytic Enzymes [J]. Biotechnology Bulletin, 2020, 36(10): 226-236. |
| [11] | WANG Qi-wen, LI Pan, Pan Cui-yun, HAN Fen-xia. Effect of Ethylene Glycol on the Expression of Exogenous Genes in Vivo [J]. Biotechnology Bulletin, 2019, 35(4): 64-68. |
| [12] | LIU Chang-yu, CHEN Xun, LONG Yu-qing, CHEN Ya, LIU Xiang-dan, ZHOU Ri-bao. Research Advances in Genes Involved in Ethylene Biosynthesis and Signal Transduction During Flower Senescence [J]. Biotechnology Bulletin, 2019, 35(3): 171-182. |
| [13] | WANG Zhu-cheng, LIU Hui, LI Rong-hua, CHEN Xin, LI Xin, LU Zhi-yuan. Physiological Mechanism of Exogenous Ethylene and Sulfur in Alleviating Cadmium Stress in Portulaca oleracea [J]. Biotechnology Bulletin, 2019, 35(10): 71-79. |
| [14] | DOU Yue, LIU Mei-tong, LU An-na, WU Jia-jie, WANG Qun-qing, XU Qian. Regulatory Mechanism of Mediator Subunit MED25 on Multi-phytohormone Signaling Pathways [J]. Biotechnology Bulletin, 2018, 34(7): 40-47. |
| [15] | YUAN Lin, HUANG Zhao, ZENG Jing, GUO Jian-jun, ZHANG Ting, Lü Jun. Fusion of Phytase YiAPPA with the Raw-starch Binding Domain and Characterization of the Fusion Enzyme [J]. Biotechnology Bulletin, 2018, 34(3): 200-207. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||