








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (5): 136-148.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0998
Previous Articles Next Articles
ZHANG Hao( ), LI Zhe(
), LI Zhe( ), GUO Kai, HUANG Yan-hua, HAO Yong-ren
), GUO Kai, HUANG Yan-hua, HAO Yong-ren
Received:2021-08-05
Online:2022-05-26
Published:2022-06-10
Contact:
LI Zhe
E-mail:arthur02@163.com;zhel@sdas.org
ZHANG Hao, LI Zhe, GUO Kai, HUANG Yan-hua, HAO Yong-ren. Functional Analysis of TvGCN5 Gene Encoding Histone Acetylase from Trichoderma viride Tv-1511[J]. Biotechnology Bulletin, 2022, 38(5): 136-148.
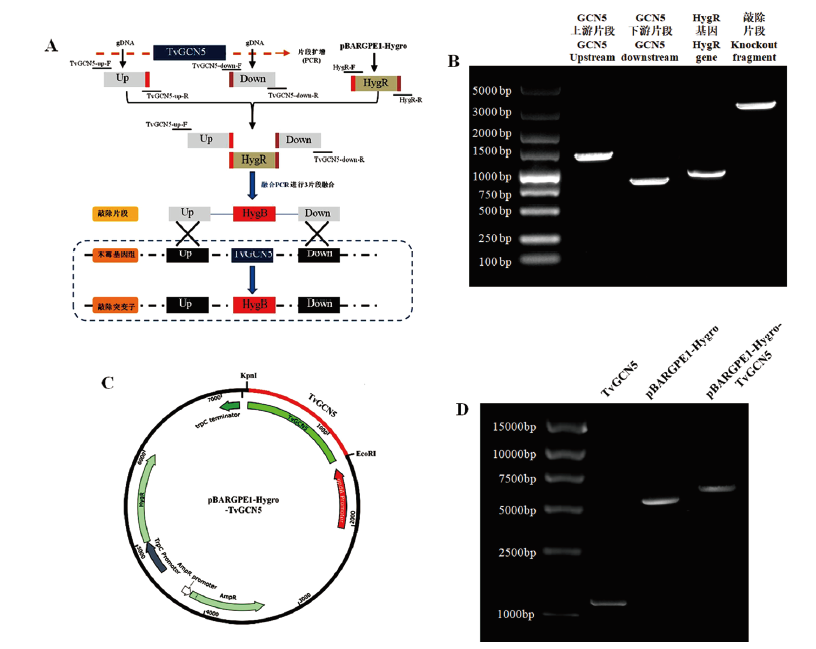
Fig. 1 Construction of knockout fragment and overexpression vector of TvGCN5 gene A,B:Fragment construction of knockout TvGCN5 gene. C,D:Construction of overexpression vector of TvGCN5 gene

Fig.2 Identification of overexpressing or deleting TvGCN5 gene in Trichoderma engineering strains by fluores-cence quantitative PCR and Western blot A:TvGCN5 gene transcription level in Trichoderma engineered strains by fluorescence quantitative PCR. B,C:Protein expression level of TvGCN5 in Trichoderma engineered strains by Western blot

Fig.3 Changes in the levels of histone acetylation in T. viride Tv-1511 and its engineered strains A:Quantitative detection of histone H3 acetylation level in T. viride Tv-1511 and its engineered strains by assay kit. B,C:Detection of histone H3 acetylation level in T. viride Tv-1511and its engineered strains by Western blot
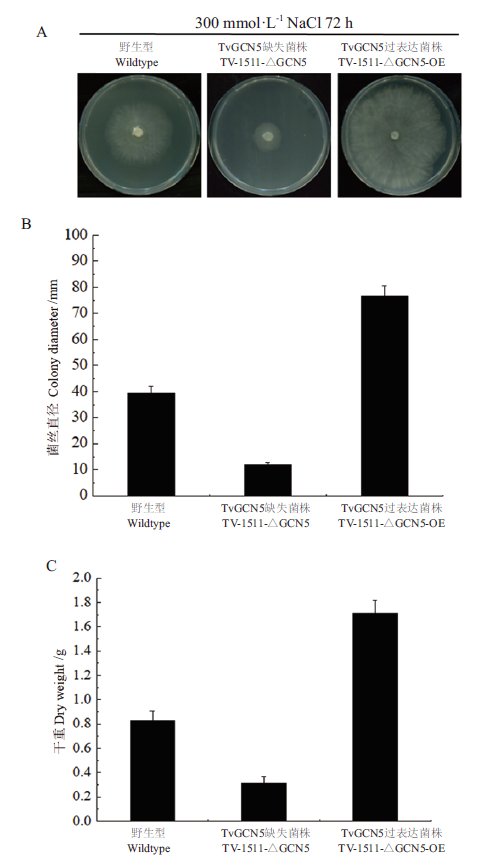
Fig.4 Analysis of the salt tolerances of T. viride Tv-1511 original strain and its engineered strains A,B:Petri dish test of the salt tolerance of Trichoderma and quantitative statistics of colony diameter. C:Dry weight of Trichoderma in liquid culture mycelium under salt stress

Fig.5 Analysis of the high temperature tolerance of T. vir-ide Tv-1511 original strain and its engineered strains A,B:Petri dish test of the heat tolerance of Trichoderma and quantitative statistics of colony diameter. C:Dry weight of Trichoderma liquid culture mycelium under high temperature stress

Fig.6 Analysis of the growth-promoting effects of T. viride Tv-1511 original strain and its engineering strains on Arabidopsis thaliana A:Promoting effects of T. viride and its engineering strains on the growth of A. thaliana. B:Quantitative statistics of fresh weight of Arabidopsis treated with T. viride and its engineered strains. C:Quantitative statistics of leaf areas of Arabidopsis treated with T. viride and its engineering strains

Fig.7 Analysis of the growth-promoting effects of T. viride Tv-1511 original strain and its engineering strains on peppermint(Mentha piperita) A:Promoting effects of Trichoderma viride Tv-1511 and its engineering strains on the growth of peppermint. B:Quantitative statistics of fresh weight of peppermint treated with Trichoderma strains. C:Quantitative statistics of leaf area of peppermint treated with Trichoderma strains
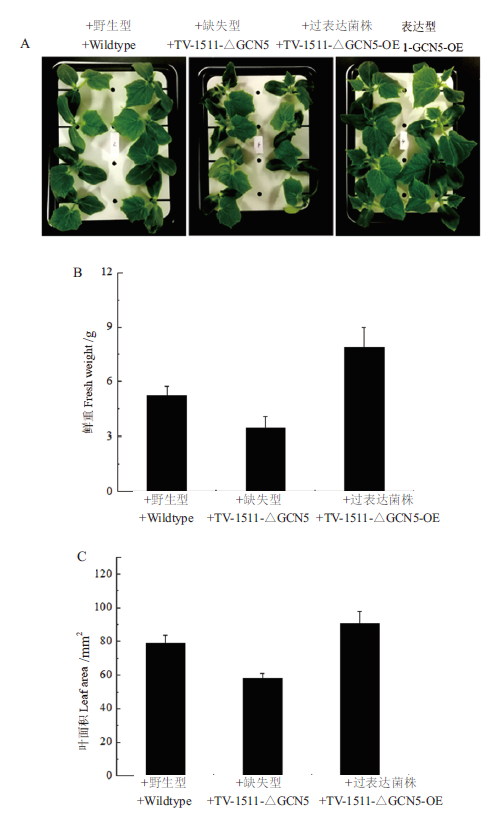
Fig.8 Analysis of the promoting effects of T. viride Tv-1511 original strain and its engineering strains on cucum-ber(Cucumis sativus L.) A:Promoting effects of Trichoderma viride Tv-1511 and its engineering strains on the growth of cucumber. B:Quantitative statistics of fresh weight of cucumber treated with Trichoderma strains. C:Quantitative statistics of leaf areas of cucumber treated with Trichoderma strains

Fig.9 Detection of IAA contents in the fermentation broths of T. viride Tv-1511 original strain and its engineer-ing strains A:Chromatogram of IAA standard. B:Quantitative statistics of IAA content in the fermentation broth of T. viride Tv-1511 and its engineering strains

Fig.10 Analysis of cellulase activities of T. viride Tv-1511 original strain and its engineering strains A:Detection of filter paper enzyme activity in the fermentation broth of T. viride Tv-1511 and its engineering strains. B:Detection of hydroxymethyl cellulase activity in the fermentation broth of T. viride Tv-1511 and its engineering strains. C:Detection of exoglucanase activity in the fermentation broth of T. viride Tv-1511 and its engineering strains. D:Detection of β-glucosidase activity in the fermentation broth of T. viride Tv-1511 and its engineering strains
| [1] | 闫峰, 徐凤花, 顾金刚, 等. 木霉属真菌的生物降解及生物转化作用研究进展[J]. 微生物学杂志, 2009, 29(3):77-80. |
| Yan F, Xu FH, Gu JG, et al. Research progress of biodegradation and biotransformation with Trichoderma[J]. J Microbiol, 2009, 29(3):77-80. | |
| [2] | 高克祥, 刘晓光, 郭润芳, 等. 木霉菌对五种植物病原真菌的重寄生作用[J]. 山东农业大学学报:自然科学版, 2002, 33(1):37-42. |
| Gao KX, Liu XG, Guo RF, et al. Mycoparasitism of Trichoderma spp. five plant pathogenic fungi[J]. J Shandong Agric Univ, 2002, 33(1):37-42. | |
| [3] |
Gajera HP, Katakpara ZA, Patel SV, et al. Antioxidant defense response induced by Trichoderma viride against Aspergillus niger Van Tieghem causing collar rot in groundnut(Arachis hypogaea L.)[J]. Microb Pathog, 2016, 91:26-34.
doi: 10.1016/j.micpath.2015.11.010 URL |
| [4] |
Saravanakumar K, Yu CJ, Dou K, et al. Synergistic effect of Trichoderma-derived antifungal metabolites and cell wall degrading enzymes on enhanced biocontrol of Fusarium oxysporum f. sp. cucumerinum[J]. Biol Control, 2016, 94:37-46.
doi: 10.1016/j.biocontrol.2015.12.001 URL |
| [5] |
Landero Valenzuela N, Nieto Angel D, Téliz Ortiz D, et al. Biological control of anthracnose by postharvest application of Trichoderma spp. on maradol Papaya fruit[J]. Biol Control, 2015, 91:88-93.
doi: 10.1016/j.biocontrol.2015.08.002 URL |
| [6] |
Hu XJ, Roberts DP, Xie LH, et al. Use of formulated Trichoderma sp. Tri-1 in combination with reduced rates of chemical pesticide for control of Sclerotinia sclerotiorium on oilseed rape[J]. Crop Prot, 2016, 79:124-127.
doi: 10.1016/j.cropro.2015.10.020 URL |
| [7] |
Arjona-Girona I, Vinale F, Ruano-Rosa D, et al. Effect of metabolites from different Trichoderma strains on the growth of Rosellinia necatrix, the causal agent of avocado white root rot[J]. Eur J Plant Pathol, 2014, 140(2):385-397.
doi: 10.1007/s10658-014-0472-z URL |
| [8] | 庄敬华, 杨长城, 牟连晓, 等. 土壤不同处理对木霉菌定殖及其生防效果的影响[J]. 植物保护, 2005, 31(6):42-44. |
| Zhuang JH, Yang CC, Mu LX, et al. Effect of soil treatment on colonization and biocontrol efficiency of Trichoderma[J]. Plant Prot, 2005, 31(6):42-44. | |
| [9] |
Doni F, Zain CRCM, Isahak A, et al. Relationships observed between Trichoderma inoculation and characteristics of rice grown under System of Rice Intensification(SRI)vs. conventional methods of cultivation[J]. Symbiosis, 2017, 72(1):45-59.
doi: 10.1007/s13199-016-0438-3 URL |
| [10] |
Harman GE, Howell CR, Viterbo A, et al. Trichoderma species—opportunistic, avirulent plant symbionts[J]. Nat Rev Microbiol, 2004, 2(1):43-56.
pmid: 15035008 |
| [11] |
Harman GE. Overview of mechanisms and uses of Trichoderma spp[J]. Phytopathology, 2006, 96(2):190-194.
doi: 10.1094/PHYTO-96-0190 pmid: 18943924 |
| [12] | Stewart A, Hill R. Applications of Trichoderma in plant growth promotion[M]// Biotechnology and Biology of Trichoderma.Amsterdam:Elsevier, 2014:415-428. |
| [13] | 张玉荣. 木霉菌对宿主植物的促生抗逆作用研究进展[J]. 农业与技术, 2013, 33(12):27. |
| Zhang YR. Advances in studies on growth promoting and resistance of Trichoderma to host plants[J]. Agric Technol, 2013, 33(12):27. | |
| [14] |
Adams P, Lynch JM, De Leij FA. Desorption of zinc by extracellularly produced metabolites of Trichoderma harzianum, Trichoderma reesei and Coriolus versicolor[J]. J Appl Microbiol, 2007, 103(6):2240-2247.
pmid: 18045407 |
| [15] | 徐文. 木霉-黄瓜互作过程中抗病信号传递途径分析[D]. 天津: 河北工业大学, 2017. |
| Xu W. Analysis of resistance signaling pathways of the interaction between Trichoderma and cucumber[D]. Tianjin: Hebei University of Technology, 2017. | |
| [16] | 张量, 张敬泽. 渐绿木霉抑菌物质的分离纯化及其对植物病原菌的抑制作用[J]. 中国农业科学, 2015, 48(5):882-888. |
| Zhang L, Zhang JZ. Isolation and purification of active compound from Trichoderma viridescens and its inhibitory activities against phytopathogens[J]. Sci Agric Sin, 2015, 48(5):882-888. | |
| [17] |
Vinale F, Sivasithamparam K, Ghisalberti EL, et al. A novel role for Trichoderma secondary metabolites in the interactions with plants[J]. Physiol Mol Plant P, 2008, 72(1-3):80-86.
doi: 10.1016/j.pmpp.2008.05.005 URL |
| [18] | 姜绪林. 绿色木霉固态发酵产纤维素酶的研究[D]. 无锡: 江南大学, 2005. |
| Jiang XL. Studies on solid fermentation of cellulase by Trichoderma viride[D]. Wuxi: Jiangnan University, 2005. | |
| [19] |
Jones PO, Vasudevan PT. Cellulose hydrolysis by immobilized Trichoderma reesei cellulase[J]. Biotechnol Lett, 2010, 32(1):103-106.
doi: 10.1007/s10529-009-0119-x URL |
| [20] | Burgess RJ, Zhang Z. Roles for Gcn5 in promoting nucleosome assembly and maintaining genome integrity[J]. Cell Cycle, 2010, 9(15):2979-2985. |
| [21] |
Carraro DM, Ferreira Júnior JR, Schumacher R, et al. A region of the cellobiohydrolase I promoter from the filamentous fungus Trichoderma reesei mediates glucose repression in Saccharomyces cerevisiae, dependent on mitochondrial activity[J]. Biochem Biophys Res Commun, 1998, 253(2):407-414.
doi: 10.1006/bbrc.1998.9758 URL |
| [22] |
Fan L, Fu K, Yu C, et al. Thc6 protein, isolated from Trichoderma harzianum, can induce maize defense response against Curvularia lunata[J]. J Basic Microbiol, 2015, 55(5):591-600.
doi: 10.1002/jobm.201300814 URL |
| [23] |
Luger K, Richmond TJ. The histone tails of the nucleosome[J]. Curr Opin Genet Dev, 1998, 8(2):140-146.
pmid: 9610403 |
| [24] |
Luger K, Mäder AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2. 8 A resolution[J]. Nature, 1997, 389(6648):251-260.
doi: 10.1038/38444 URL |
| [25] |
Brownell JE, Zhou J, Ranalli T, et al. Tetrahymena histone acetyltransferase A:a homolog to yeast Gcn5p linking histone acetylation to gene activation[J]. Cell, 1996, 84(6):843-851.
pmid: 8601308 |
| [26] |
Jiang JF, Lu JY, Lu D, et al. Investigation of the acetylation mechanism by GCN5 histone acetyltransferase[J]. PLoS One, 2012, 7(5):e36660.
doi: 10.1371/journal.pone.0036660 URL |
| [27] |
Hinnebusch AG, Fink GR. Positive regulation in the general amino acid control of Saccharomyces cerevisiae[J]. Proc Natl Acad Sci USA, 1983, 80(17):5374-5378.
doi: 10.1073/pnas.80.17.5374 URL |
| [28] |
Xin Q, Gong Y, Lv X, et al. Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, conidiation, and cellulase gene expression[J]. Curr Microbiol, 2013, 67(5):580-589.
doi: 10.1007/s00284-013-0396-4 pmid: 23748966 |
| [29] |
Zheng F, Cao Y, Lv X, et al. A copper-responsive promoter replacement system to investigate gene functions in Trichoderma reesei:a case study in characterizing SAGA genes[J]. Appl Microbiol Biotechnol, 2017, 101(5):2067-2078.
doi: 10.1007/s00253-016-8036-0 URL |
| [30] | 唐永庆, 许艳丽, 张红骥, 等. 木霉制剂的生防应用研究及发展前景[J]. 黑龙江农业科学, 2008(1):111-113. |
| Tang YQ, Xu YL, Zhang HJ, et al. Application study and development prospect of biocontrol agents of Trichoderma[J]. Heilongjiang Agric Sci, 2008(1):111-113. | |
| [31] |
Bae H, Sicher RC, Kim MS, et al. The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao[J]. J Exp Bot, 2009, 60(11):3279-3295.
doi: 10.1093/jxb/erp165 URL |
| [32] |
Jenuwein T, Allis CD. Translating the histone code[J]. Science, 2001, 293(5532):1074-1080.
pmid: 11498575 |
| [33] |
Minerdi D, Bossi S, Gullino ML, et al. Volatile organic compounds:a potential direct long-distance mechanism for antagonistic action of Fusarium oxysporum strain MSA 35[J]. Environ Microbiol, 2009, 11(4):844-854.
doi: 10.1111/j.1462-2920.2008.01805.x URL |
| [34] |
Ryu CM, Farag MA, Hu CH, et al. Bacterial volatiles promote growth in Arabidopsis[J]. Proc Natl Acad Sci USA, 2003, 100(8):4927-4932.
doi: 10.1073/pnas.0730845100 URL |
| [35] |
Kishimoto K, Matsui K, Ozawa R, et al. Volatile 1-octen-3-ol induces a defensive response in Arabidopsis thaliana[J]. J Gen Plant Pathol, 2007, 73(1):35-37.
doi: 10.1007/s10327-006-0314-8 URL |
| [36] |
Stoppacher N, Kluger B, Zeilinger S, et al. Identification and profiling of volatile metabolites of the biocontrol fungus Trichoderma atroviride by HS-SPME-GC-MS[J]. J Microbiol Methods, 2010, 81(2):187-193.
doi: 10.1016/j.mimet.2010.03.011 pmid: 20302890 |
| [37] |
Contreras-Cornejo HA, Macías-Rodríguez L, Cortés-Penagos C, et al. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis[J]. Plant Physiol, 2009, 149(3):1579-1592.
doi: 10.1104/pp.108.130369 pmid: 19176721 |
| [38] |
Li S, Lin YCJ, Wang PY, et al. Histone acetylation cooperating with AREB1 transcription factor regulates drought response and tolerance in Populus trichocarpa[J]. Plant Cell, 2019, 31(3):663-686.
doi: 10.1105/tpc.18.00437 URL |
| [39] |
Zheng M, Liu X, Lin J, et al. Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes[J]. Plant J, 2019, 97(3):587-602.
doi: 10.1111/tpj.14144 |
| [40] |
Hu Z, Song N, Zheng M, et al. Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis[J]. Plant J, 2015, 84(6):1178-1191.
doi: 10.1111/tpj.13076 URL |
| [41] | 周娇娇. 组蛋白修饰对里氏木霉纤维素酶表达的初步研究[D]. 深圳: 深圳大学, 2017. |
| Zhou JJ. Preliminary study of histone modification on cellulase expression in Trichoderma reesei[D]. Shenzhen: Shenzhen University, 2017. |
| [1] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [2] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [3] | HU Ming-yue, YANG Yu, GUO Yang-dong, ZHANG Xi-chun. Functional Analysis of SlMYB96 Gene in Tomato Under Cold Stress [J]. Biotechnology Bulletin, 2023, 39(4): 236-245. |
| [4] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [5] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [6] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [7] | LAI Xin-tong, WANG Ke-lan, YOU Yu-xin, TAN Jun-jie. Recent Advances in CRISPR/Cas-based DNA Base Editing [J]. Biotechnology Bulletin, 2022, 38(6): 1-12. |
| [8] | CHEN Ying-dan, ZHANG Yang, XIA Qiang, SUN Hong-xia. Gene Editing Technology of CRISPR/Cas and Its Applications in Microalgae Research [J]. Biotechnology Bulletin, 2022, 38(5): 257-268. |
| [9] | ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9 [J]. Biotechnology Bulletin, 2022, 38(10): 216-225. |
| [10] | LIU Sha-yu, CAO Jian, LI Meng, LIU Zhi-qiang, LI Xiao-yu. Biological Function of a Zn2Cys6 Transcription Factor CgAswA in Colletotrichum gloeosporioides [J]. Biotechnology Bulletin, 2021, 37(9): 161-170. |
| [11] | HU Xiu-wen, LIU Hua, WANG Yu, TANG Xue-ming, WANG Jin-bin, ZENG Hai-juan, JIANG Wei, LI Hong. Application of CRISPR-Cas System in Nucleic Acid Detection [J]. Biotechnology Bulletin, 2021, 37(9): 266-273. |
| [12] | HUANG Yao-hui, JIAO Yue, FU Zhong-wen. Overview and Progress of Japan Safety Management System of Genetically Modified Crops [J]. Biotechnology Bulletin, 2021, 37(3): 99-106. |
| [13] | ZUO Ling-li, ZHOU Li-ting, WU Xing-qi, WU Chao-yi, WU Shu-yan. Construction of spvBC Gene Editing Strains of Salmonella typhimurium [J]. Biotechnology Bulletin, 2021, 37(2): 253-260. |
| [14] | WANG Kai-kai, WANG Xiao-lu, SU Xiao-yun, ZHANG Jie. Optimization and Application of Double-plasmid CRISPR-Cas9 System in Escherichia coli [J]. Biotechnology Bulletin, 2021, 37(12): 252-264. |
| [15] | LIU Jia, WEI Jia-qi, LIU Yu-qin, SHI Ge-ge, GUO Jing. Research on Evolution of Gene Editing Technology Based on Patent Analysis and Social Network Analysis [J]. Biotechnology Bulletin, 2021, 37(12): 274-284. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||