








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (6): 103-111.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1006
Previous Articles Next Articles
LI Yi-han( ), YU Lang-liu, LI Chun-yan, ZHANG Meng-meng, ZHANG Xiao-qin, FANG Yun-xia(
), YU Lang-liu, LI Chun-yan, ZHANG Meng-meng, ZHANG Xiao-qin, FANG Yun-xia( ), XUE Da-wei(
), XUE Da-wei( )
)
Received:2021-07-21
Online:2022-06-26
Published:2022-07-11
Contact:
FANG Yun-xia,XUE Da-wei
E-mail:3340043951@qq.com;yxfang12@163.com;dwxue@hznu.edu.cn
LI Yi-han, YU Lang-liu, LI Chun-yan, ZHANG Meng-meng, ZHANG Xiao-qin, FANG Yun-xia, XUE Da-wei. Whole Genome Identification of Barley NRAMP and Gene Expression Analysis Under Heavy Metal Stress[J]. Biotechnology Bulletin, 2022, 38(6): 103-111.
| 基因 Gene | 基因ID Gene ID | 引物 Primer | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|---|---|
| HvNramp1 | HORVU3Hr1G042440 | HvqRT1 | CTACCTTCGTGAGTTGGCTTGTA | TGTTGTCTTGTATTGGCACCTCT |
| HvNramp2 | HORVU4Hr1G013220 | HvqRT2 | AGCACCATAACCGGGACCTA | TGAGCCACTCGTTGAGGACAT |
| HvNramp3 | HORVU4Hr1G069320 | HvqRT3 | TGTTTACCTCGCCTTCATCGT | CTACCCACTTCCGCTCATCTT |
| HvNramp4 | HORVU4Hr1G077510 | HvqRT4 | AGGACCTCGCAGACATACCG | AGAAGGACGGATAAAATGCTACG |
| HvNramp5 | HORVU4Hr1G090460 | HvqRT5 | GGTCTCCAAGCTGGGATGTCT | ATGGCTGTGATAAAATCATCCTG |
| HvNramp6 | HORVU5Hr1G018850 | HvqRT6 | AACCGAGCCGTACAATGGAC | AGATGATCGTGGCGATACGC |
| HvNramp7 | HORVU5Hr1G050310 | HvqRT7 | CTGCATAGCAGGTCTCGCACT | CATTTCCACCCAGAAGTGCC |
| HvNramp8 | HORVU7Hr1G078330 | HvqRT8 | GTGAGGTGAACACTGGCCAGA | CCACCATTATGCCTAAAGAGCC |
| HvNramp9 | HORVU5Hr1G050330 | HvqRT9 | ATCGGGATAAACGTCTACTTCCT | ATGACAGAGGCGACGTAGAGC |
| HvNramp10 | HORVU7Hr1G106570 | HvqRT10 | GTGCTAACATGGGCGATCG | ACGGCAAGGTACACTTCCTGTT |
| HvActin | HORVU1Hr1G002840 | Actin | TGGATCGGAGGGTCCATCCT | GCACTTCCTGTGGACGATCGCTG |
Table 1 Primers for qRT- PCR analysis
| 基因 Gene | 基因ID Gene ID | 引物 Primer | 正向引物序列 Forward primer sequence(5'-3') | 反向引物序列 Reverse primer sequence(5'-3') |
|---|---|---|---|---|
| HvNramp1 | HORVU3Hr1G042440 | HvqRT1 | CTACCTTCGTGAGTTGGCTTGTA | TGTTGTCTTGTATTGGCACCTCT |
| HvNramp2 | HORVU4Hr1G013220 | HvqRT2 | AGCACCATAACCGGGACCTA | TGAGCCACTCGTTGAGGACAT |
| HvNramp3 | HORVU4Hr1G069320 | HvqRT3 | TGTTTACCTCGCCTTCATCGT | CTACCCACTTCCGCTCATCTT |
| HvNramp4 | HORVU4Hr1G077510 | HvqRT4 | AGGACCTCGCAGACATACCG | AGAAGGACGGATAAAATGCTACG |
| HvNramp5 | HORVU4Hr1G090460 | HvqRT5 | GGTCTCCAAGCTGGGATGTCT | ATGGCTGTGATAAAATCATCCTG |
| HvNramp6 | HORVU5Hr1G018850 | HvqRT6 | AACCGAGCCGTACAATGGAC | AGATGATCGTGGCGATACGC |
| HvNramp7 | HORVU5Hr1G050310 | HvqRT7 | CTGCATAGCAGGTCTCGCACT | CATTTCCACCCAGAAGTGCC |
| HvNramp8 | HORVU7Hr1G078330 | HvqRT8 | GTGAGGTGAACACTGGCCAGA | CCACCATTATGCCTAAAGAGCC |
| HvNramp9 | HORVU5Hr1G050330 | HvqRT9 | ATCGGGATAAACGTCTACTTCCT | ATGACAGAGGCGACGTAGAGC |
| HvNramp10 | HORVU7Hr1G106570 | HvqRT10 | GTGCTAACATGGGCGATCG | ACGGCAAGGTACACTTCCTGTT |
| HvActin | HORVU1Hr1G002840 | Actin | TGGATCGGAGGGTCCATCCT | GCACTTCCTGTGGACGATCGCTG |
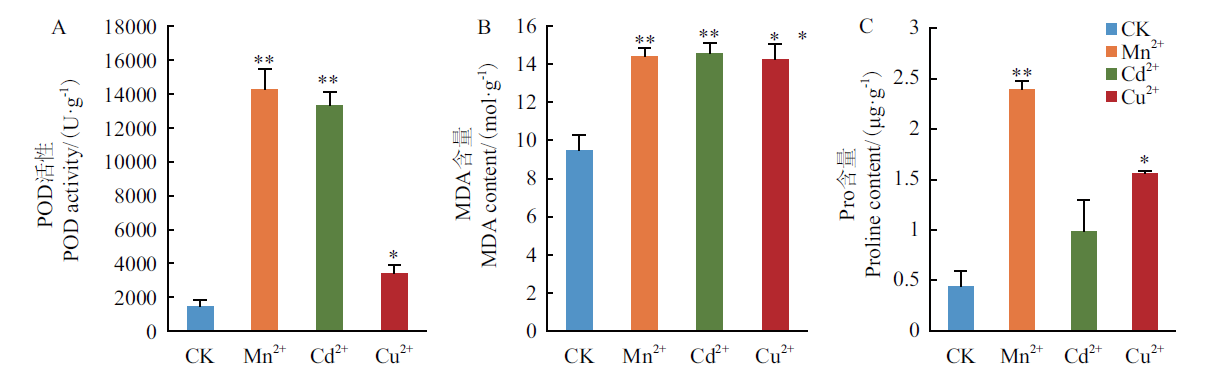
Fig. 1 Effects of heavy metal ion stress on the physiological indexes of barley seedling leaves * and ** indicate significant or extremely significant differences between the heavy metal ion stress treatment and the control group,respectively. The independent sample t-test(*P<0.05,**P<0.01)was used for differences analysis,and error bars indicate standard errors
| 基因 Gene | 蛋白质 Protein | 氨基酸长度 Amino acid length/aa | 分子质量 Molecular mass/Da | 理论等电点 Theoretical isoelectric point | 亚细胞定位 Subcellular localization | 跨膜结构域 Transmembrane domain | 染色体定位 Chromosomal localization | 物理图谱位置 Physical map location/Mb |
|---|---|---|---|---|---|---|---|---|
| HvNramp1 | A0A287KVZ6 | 606 | 66 306.78 | 7.82 | 细胞膜Cell membrane | 12 | Chr.3H | 256.8 |
| HvNramp2 | A0A287N9P9 | 549 | 60 120.01 | 5.07 | 细胞膜Cell membrane | 10 | Chr.4H | 43.2 |
| HvNramp3 | M0XUV1 | 522 | 56 885.5 | 5.76 | 细胞膜Cell membrane | 10 | Chr.4H | 543.5 |
| HvNramp4 | M0X4N6 | 542 | 58 612.97 | 7.03 | 细胞膜Cell membrane | 11 | Chr.4H | 576.7 |
| HvNramp5 | A0A287Q4H3 | 212 | 22 845.85 | 5.17 | 细胞膜Cell membrane | 5 | Chr.4H | 615.3 |
| HvNramp6 | F2EHM2 | 547 | 58 995.65 | 5.2 | 细胞膜Cell membrane | 11 | Chr.5H | 75.1 |
| HvNramp7 | A0A287R642 | 248 | 58 995.65 | 5.41 | 细胞膜Cell membrane | 7 | Chr.5H | 373.2 |
| HvNramp8 | A0A287X031 | 515 | 55 875.86 | 8.12 | 细胞核Nucleus | 5 | Chr.7H | 438.5 |
| HvNramp9 | A0A287R6C2 | 1 010 | 110 400.77 | 6.13 | 细胞膜Cell membrane | 11 | Chr.5H | 373.2 |
| HvNramp10 | M0W7C8 | 548 | 59 571.99 | 7.99 | 细胞膜Cell membrane | 12 | Chr.7H | 591.4 |
Table 2 Physicochemical properties and chromosome location of NRAMP protein in barley
| 基因 Gene | 蛋白质 Protein | 氨基酸长度 Amino acid length/aa | 分子质量 Molecular mass/Da | 理论等电点 Theoretical isoelectric point | 亚细胞定位 Subcellular localization | 跨膜结构域 Transmembrane domain | 染色体定位 Chromosomal localization | 物理图谱位置 Physical map location/Mb |
|---|---|---|---|---|---|---|---|---|
| HvNramp1 | A0A287KVZ6 | 606 | 66 306.78 | 7.82 | 细胞膜Cell membrane | 12 | Chr.3H | 256.8 |
| HvNramp2 | A0A287N9P9 | 549 | 60 120.01 | 5.07 | 细胞膜Cell membrane | 10 | Chr.4H | 43.2 |
| HvNramp3 | M0XUV1 | 522 | 56 885.5 | 5.76 | 细胞膜Cell membrane | 10 | Chr.4H | 543.5 |
| HvNramp4 | M0X4N6 | 542 | 58 612.97 | 7.03 | 细胞膜Cell membrane | 11 | Chr.4H | 576.7 |
| HvNramp5 | A0A287Q4H3 | 212 | 22 845.85 | 5.17 | 细胞膜Cell membrane | 5 | Chr.4H | 615.3 |
| HvNramp6 | F2EHM2 | 547 | 58 995.65 | 5.2 | 细胞膜Cell membrane | 11 | Chr.5H | 75.1 |
| HvNramp7 | A0A287R642 | 248 | 58 995.65 | 5.41 | 细胞膜Cell membrane | 7 | Chr.5H | 373.2 |
| HvNramp8 | A0A287X031 | 515 | 55 875.86 | 8.12 | 细胞核Nucleus | 5 | Chr.7H | 438.5 |
| HvNramp9 | A0A287R6C2 | 1 010 | 110 400.77 | 6.13 | 细胞膜Cell membrane | 11 | Chr.5H | 373.2 |
| HvNramp10 | M0W7C8 | 548 | 59 571.99 | 7.99 | 细胞膜Cell membrane | 12 | Chr.7H | 591.4 |
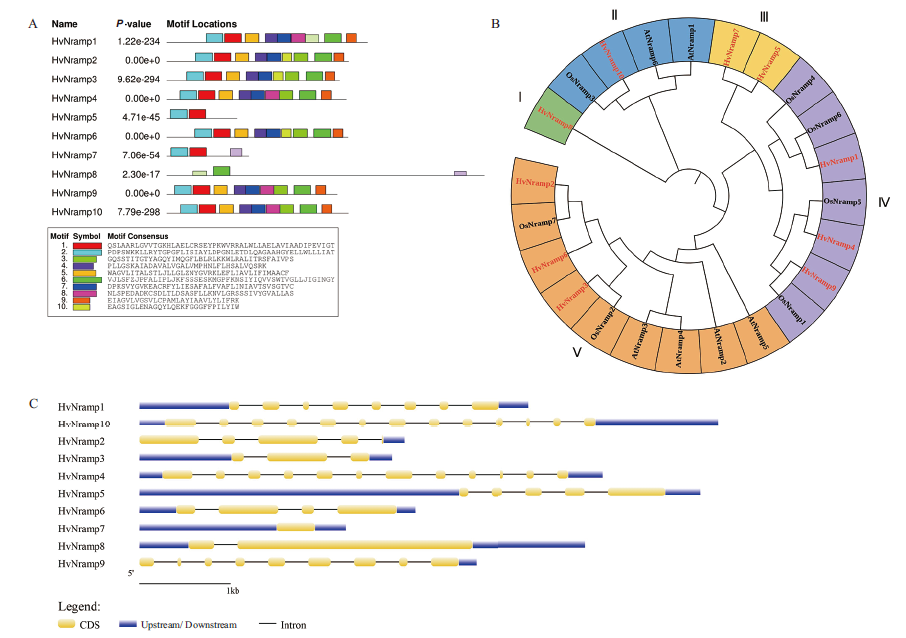
Fig. 2 Conserved sequences,gene structure and phylogenetic analysis of barley NRAMP gene family A:Barley NRAMP family protein conservative sequence distribution. B:Barley,rice and Arabidopsis NRAMP gene family system evolution tree. Barley NRAMP system evolution analysis,different color branches represent different subfamilies(I-V),red labeled as HvNramp. C:Intron and exon structure of barley NRAMP gene
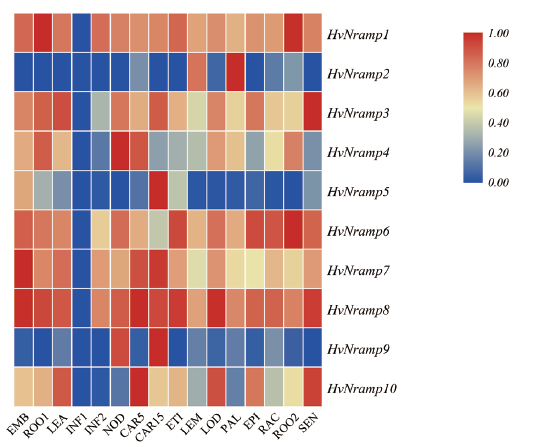
Fig. 3 Expression profile analysis of NRAMP gene family in barley EMB:Embryonic tissue. ROO1:10 cm root tissue. LEA:Leaf tissue. INF2:Inflorescence tissue. NOD:Internode of the third stem. CAR5:Caryopsis 5 d after flowering. CAR15:Caryopsis 15 d after flowering. ETI:Etching leaves. LEM:Inflorescence lemma. LOD:Inflorescence lodicule. PAL:Inflorescence lemma. EPI:Striped epidermis. RAC:Inflorescence rachis. ROO2:28 d root tissue. SEN:Old leaves
| [1] | 李洋, 于丽杰, 金晓霞. 植物重金属胁迫耐受机制[J]. 中国生物工程杂志, 2015, 35(9):94-104. |
| Li Y, Yu LJ, Jin XX. Mechanism of heavy metal tolerance stress of plants[J]. China Biotechnol, 2015, 35(9):94-104. | |
| [2] |
Fang Y, Deng X, Lu X, et al. Differential phosphoproteome study of the response to cadmium stress in rice[J]. Ecotoxicol Environ Saf, 2019, 180:780-788.
doi: 10.1016/j.ecoenv.2019.05.068 URL |
| [3] | 吕夏晨, 徐玲, 张蓝天, 等. 褪黑素对干旱胁迫下大麦生理及蜡质基因表达的影响[J]. 植物生理学报, 2020, 56(5):1073-1080. |
| Lü XC, Xu L, Zhang LT, et al. Effects of exogenous melatonin on physiology and waxy genes expression in barley under drought stress[J]. Plant Physiol J, 2020, 56(5):1073-1080. | |
| [4] |
Skamene E, Pietrangeli CE. Genetics of the immune response to infectious pathogens[J]. Curr Opin Immunol, 1991, 3(4):511-517.
pmid: 1755977 |
| [5] | 陈可欣, 蒋贤达, 朱祝军, 等. 植物Nramp家族参与金属离子吸收和分配的研究进展[J]. 植物生理学报, 2020, 56(3):345-355. |
|
Chen KX, Jiang XD, Zhu ZJ, et al. Advances in the study of plant Nramp family involved in metal ion absorption and distribution[J]. Plant Physiol J, 2020, 56(3):345-355.
doi: 10.1104/pp.56.3.345 URL |
|
| [6] |
Peris-Peris C, Serra-Cardona A, Sánchez-Sanuy F, et al. Two NRAMP6 isoforms function as iron and manganese transporters and contribute to disease resistance in rice[J]. Mol Plant Microbe Interact, 2017, 30(5):385-398.
doi: 10.1094/MPMI-01-17-0005-R URL |
| [7] |
Cailliatte R, Schikora A, Briat JF, et al. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions[J]. Plant Cell, 2010, 22(3):904-917.
doi: 10.1105/tpc.109.073023 URL |
| [8] |
Takahashi R, Ishimaru Y, Senoura T, et al. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice[J]. J Exp Bot, 2011, 62(14):4843-4850.
doi: 10.1093/jxb/err136 pmid: 21697258 |
| [9] |
Mäser P, Thomine S, Schroeder JI, et al. Phylogenetic relationships within cation transporter families of Arabidopsis[J]. Plant Physiol, 2001, 126(4):1646-1667.
pmid: 11500563 |
| [10] | 胡润芳, 林国强, 张广庆, 等. 大豆Nramp基因家族的序列特征与亲缘关系分析[J]. 分子植物育种, 2011, 9(2):245-250. |
| Hu RF, Lin GQ, Zhang GQ, et al. Analysis on soybean Nramp gene family sequence features and phylogeny[J]. Mol Plant Breed, 2011, 9(2):245-250. | |
| [11] | 卢婷婷, 李艳玲, 李存法, 等. 甘草NRAMP基因家族的鉴定和生物信息学分析[J]. 河南农业科学, 2019, 48(2):40-47. |
| Lu TT, Li YL, Li CF, et al. Identification and bioinformatics analysis of NRAMP gene family in Glycyrrhiza uralensis[J]. J Henan Agric Sci, 2019, 48(2):40-47. | |
| [12] |
Zhang XQ, Tong T, Tian B, et al. Physiological, biochemical and molecular responses of barley seedlings to aluminum stress[J]. Phyton, 2019, 88(3):253-260.
doi: 10.32604/phyton.2019.06143 URL |
| [13] | 徐晓阳, 李国龙, 孙亚卿, 等. 甜菜NAC转录因子鉴定及其在水分胁迫下的表达分析[J]. 植物生理学报, 2019, 55(4):444-456. |
| Xu XY, Li GL, Sun YQ, et al. Identification of NAC transcription factors in sugar beet and their expression analyses under water stress[J]. Plant Physiol J, 2019, 55(4):444-456. | |
| [14] |
Mayer KF, Waugh R, Brown JW, et al. A physical, genetic and functional sequence assembly of the barley genome[J]. Nature, 2012, 491(7426):711-716.
doi: 10.1038/nature11543 URL |
| [15] |
Zhang X, Zhang LT, Chen YY, et al. Genome-wide identification of the SOD gene family and expression analysis under drought and salt stress in barley[J]. Plant Growth Regul, 2021, 94(1):49-60.
doi: 10.1007/s10725-021-00695-8 URL |
| [16] |
Mani A, Sankaranarayanan K. In silico analysis of natural resistance-associated macrophage protein(NRAMP)family of transporters in rice[J]. Protein J, 2018, 37(3):237-247.
doi: 10.1007/s10930-018-9773-y URL |
| [17] | 王利华. 拟南芥AtNRAMP3转运蛋白的结构与功能研究[D]. 南京: 南京农业大学, 2017:1-74. |
| Wang LH.Structure and function of AtNRAMP 3 transporter in Arabidopsis thaliana[D]. Nanjing: Nanjing Agricultural University, 2017:1-74. | |
| [18] | 胡阳阳, 孙琳琳, 王多祥, 等. 水稻NRL基因家族的全基因组鉴定与生物信息学分析[J]. 分子植物育种, 2020. https://kns.cnki.net/kcms/detail/46.1068.S.20200904.1146.006.html. |
| Hu YY, Sun LL, Wang DX, et al. Genome-wide identification and bioinformatics analysis of NRL gene family in rice[J]. Mol Plant Breed, 2020. https://kns.cnki.net/kcms/detail/46.1068.S.20200904.1146.006.html. | |
| [19] | 杨猛. 水稻NRAMP家族基因在Mn和Cd转运中的功能研究[D]. 武汉: 华中农业大学, 2014:1-139. |
| Yang M. Functional analysis of rice NRAMP genes in Mn and Cd transport[D]. Wuhan: Huazhong Agricultural University, 2014:1-139. | |
| [20] |
Zhang XQ, Chen HN, Jiang H, et al. Measuring the damage of heavy metal cadmium in rice seedlings by SRAP analysis combined with physiological and biochemical parameters[J]. J Sci Food Agric, 2015, 95(11):2292-2298.
doi: 10.1002/jsfa.6949 URL |
| [21] | Yuchun RAO, Ran JIAO, Sheng WANG, et al. SPL36 encodes a receptor-like protein kinase that regulates programmed cell death and defense responses in rice[J]. Rice:N Y, 2021, 14(1):34. |
| [22] | 何凤, 刘攀峰, 王璐, 等. 干旱胁迫及复水对杜仲苗生理特性的影响[J]. 植物生理学报, 2021, 57(3):661-671. |
| He F, Liu PF, Wang L, et al. Effect of drought stress and rewatering on physiological characteristics of Eucommia ulmoides seedling[J]. Plant Physiol J, 2021, 57(3):661-671. | |
| [23] |
Bairoch A. The PROSITE dictionary of sites and patterns in proteins, its current status[J]. Nucleic Acids Res, 1993, 21(13):3097-3103.
pmid: 8332530 |
| [24] |
刘营, 尹泽, 江姚兰, 等. 甘蔗ScNRAMP基因家族的全基因组鉴定与生物信息学分析[J/OL]. 广西植物, 2021. DOI: 10.11931/guihaia.gxzw202104005.
doi: 10.11931/guihaia.gxzw202104005 |
|
Liu Y, Yin Z, Jiang YL, et al. Identification and bioinformatics analysis of ScNRAMP gene family in sugarcane[J/OL]. Guihaia, 2021. DOI: 10.11931/guihaia.gxzw202104005.
doi: 10.11931/guihaia.gxzw202104005 |
|
| [25] |
Gao H, Xie W, Yang C, et al. NRAMP2, a trans-Golgi network-localized manganese transporter, is required for Arabidopsis root growth under manganese deficiency[J]. New Phytol, 2018, 217(1):179-193.
doi: 10.1111/nph.14783 URL |
| [26] |
Lanquar V, Ramos MS, Lelièvre F, et al. Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency[J]. Plant Physiol, 2010, 152(4):1986-1999.
doi: 10.1104/pp.109.150946 URL |
| [27] |
Thomine S, Wang R, Ward JM, et al. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes[J]. PNAS, 2000, 97(9):4991-4996.
pmid: 10781110 |
| [28] |
Cellier M, Govoni G, Vidal S, et al. Human natural resistance-associated macrophage protein:cDNA cloning, chromosomal mapping, genomic organization, and tissue-specific expression[J]. J Exp Med, 1994, 180(5):1741-1752.
doi: 10.1084/jem.180.5.1741 pmid: 7964458 |
| [29] |
Ishimaru Y, Bashir K, Nakanishi H, et al. OsNRAMP5, a major player for constitutive iron and manganese uptake in rice[J]. Plant Signal Behav, 2012, 7(7):763-766.
doi: 10.4161/psb.20510 URL |
| [30] | 李亚敏, 巩宗强, 贾春云, 等. 玉米镉转运基因ZmNramp1的鉴定及对镉胁迫的响应[J]. 生态学杂志, 2021, 40(7):2016-2023. |
| Li YM, Gong ZQ, Jia CY, et al. Identification of the cadmium transport gene ZmNramp1 in maize and its response to cadmium stress[J]. Chin J Ecol, 2021, 40(7):2016-2023. | |
| [31] | 孙丽娟, 程龙军. 缺Cu2+和Zn2+对水稻OsNRAMP家族基因表达与主要金属离子吸收的影响[J]. 植物生理学报, 2011, 47(9):879-884. |
| Sun LJ, Cheng LJ. Effects of deficiency of copper and zinc on OsNRAMPs expression and main metal-ions uptake in Oryza sativa L[J]. Plant Physiol J, 2011, 47(9):879-884. |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [3] | JIANG Run-hai, JIANG Ran-ran, ZHU Cheng-qiang, HOU Xiu-li. Research Progress in Mechanisms of Microbial-enhanced Phytoremediation for Lead-contaminated Soil [J]. Biotechnology Bulletin, 2023, 39(8): 114-125. |
| [4] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [5] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [6] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [7] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [8] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [9] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| [10] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [11] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| [12] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| [13] | DUAN Min-jie, LI Yi-fei, YANG Xiao-miao, WANG Chun-ping, HUANG Qi-zhong, HUANG Ren-zhong, ZHANG Shi-cai. Identification of Zinc Finger Protein DnaJ-Like Gene Family in Capsicum annuum and Its Expression Analysis Responses to High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(1): 187-198. |
| [14] | YUAN Xing, GUO Cai-hua, LIU Jin-ming, KANG Chao, QUAN Shao-wen, NIU Jian-xin. Genome-wide Identification of CONSTANS-Like Family Genes and Expression Analysis in Wanlut [J]. Biotechnology Bulletin, 2022, 38(9): 167-179. |
| [15] | GUO Bin-hui, SONG Li. Transcription of Ethylene Biosynthesis and Signaling Associated Genes in Response to Heterodera glycine Infection [J]. Biotechnology Bulletin, 2022, 38(8): 150-158. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||