








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (7): 70-79.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1231
Previous Articles Next Articles
YANG Xin-ran1( ), WANG Jian-fang1, MA Xin-hao1, ZAN Lin-sen1,2(
), WANG Jian-fang1, MA Xin-hao1, ZAN Lin-sen1,2( )
)
Received:2021-09-25
Online:2022-07-26
Published:2022-08-09
Contact:
ZAN Lin-sen
E-mail:yangxinran93@nwafu.edu.cn;zanlinsen@163.com
YANG Xin-ran, WANG Jian-fang, MA Xin-hao, ZAN Lin-sen. Expression Analyses of m6A Methylase Genes in Bovine Adipogenesis[J]. Biotechnology Bulletin, 2022, 38(7): 70-79.
| 基因名称 Gene name | 引物序列 Primer sequence(5'-3') | 产物长度 Product length/bp |
|---|---|---|
| METTL3 | F:TCGAAAGCTGCACTTCAGAC | 199 |
| R:TCCAACGCTCTGTGTAAGGG | ||
| METTL14 | F:TGACATCAGAGAACTGACACCC | 198 |
| R:AGGTCCAATCCTTCCCCAGA | ||
| WTAP | F:GCCTGGAAGTTTACGCCTGA | 176 |
| R:TCCTGACTGCTTTTAAGCTCCT | ||
| FTO | F:AGCAGCGTACAACGTCACTT | 193 |
| R:AGGGTCGTCCTCACTTTCCT | ||
| ALKBH5 | F:TACTTCTTCGGCGAGGGCTA | 191 |
| R:TGGTAGTCGTTGATGACGGC | ||
| PPARγ | F:TGAAGAGCCTTCCAACTCCC | 117 |
| R:GTCCTCCGGAAGAAACCCTTG | ||
| C/EBPα | F:ATCTGCGAACACGAGACG | 73 |
| R:CCAGGAACTCGTCGTTGAA | ||
| C/EBPβ | F:TTCCTCTCCGACCTCTTCTC | 79 |
| R:CCAGACTCACGTAGCCGTACT | ||
| FABP4 | F:TGAGATTTCCTTCAAATTGGG | 101 |
| R:CTTGTACCAGAGCACCTTCATC | ||
| GAPDH | F:AGTTCAACGGCACAGTCAAGG | 124 |
| R:ACCACATACTCAGCACCAGCA | ||
| β-actin | F:CATCGGCAATGAGCGGTTCC | 147 |
| R:ACCGTGTTGGCGTAGAGGTC |
Table 1 Primers for RT-qPCR
| 基因名称 Gene name | 引物序列 Primer sequence(5'-3') | 产物长度 Product length/bp |
|---|---|---|
| METTL3 | F:TCGAAAGCTGCACTTCAGAC | 199 |
| R:TCCAACGCTCTGTGTAAGGG | ||
| METTL14 | F:TGACATCAGAGAACTGACACCC | 198 |
| R:AGGTCCAATCCTTCCCCAGA | ||
| WTAP | F:GCCTGGAAGTTTACGCCTGA | 176 |
| R:TCCTGACTGCTTTTAAGCTCCT | ||
| FTO | F:AGCAGCGTACAACGTCACTT | 193 |
| R:AGGGTCGTCCTCACTTTCCT | ||
| ALKBH5 | F:TACTTCTTCGGCGAGGGCTA | 191 |
| R:TGGTAGTCGTTGATGACGGC | ||
| PPARγ | F:TGAAGAGCCTTCCAACTCCC | 117 |
| R:GTCCTCCGGAAGAAACCCTTG | ||
| C/EBPα | F:ATCTGCGAACACGAGACG | 73 |
| R:CCAGGAACTCGTCGTTGAA | ||
| C/EBPβ | F:TTCCTCTCCGACCTCTTCTC | 79 |
| R:CCAGACTCACGTAGCCGTACT | ||
| FABP4 | F:TGAGATTTCCTTCAAATTGGG | 101 |
| R:CTTGTACCAGAGCACCTTCATC | ||
| GAPDH | F:AGTTCAACGGCACAGTCAAGG | 124 |
| R:ACCACATACTCAGCACCAGCA | ||
| β-actin | F:CATCGGCAATGAGCGGTTCC | 147 |
| R:ACCGTGTTGGCGTAGAGGTC |
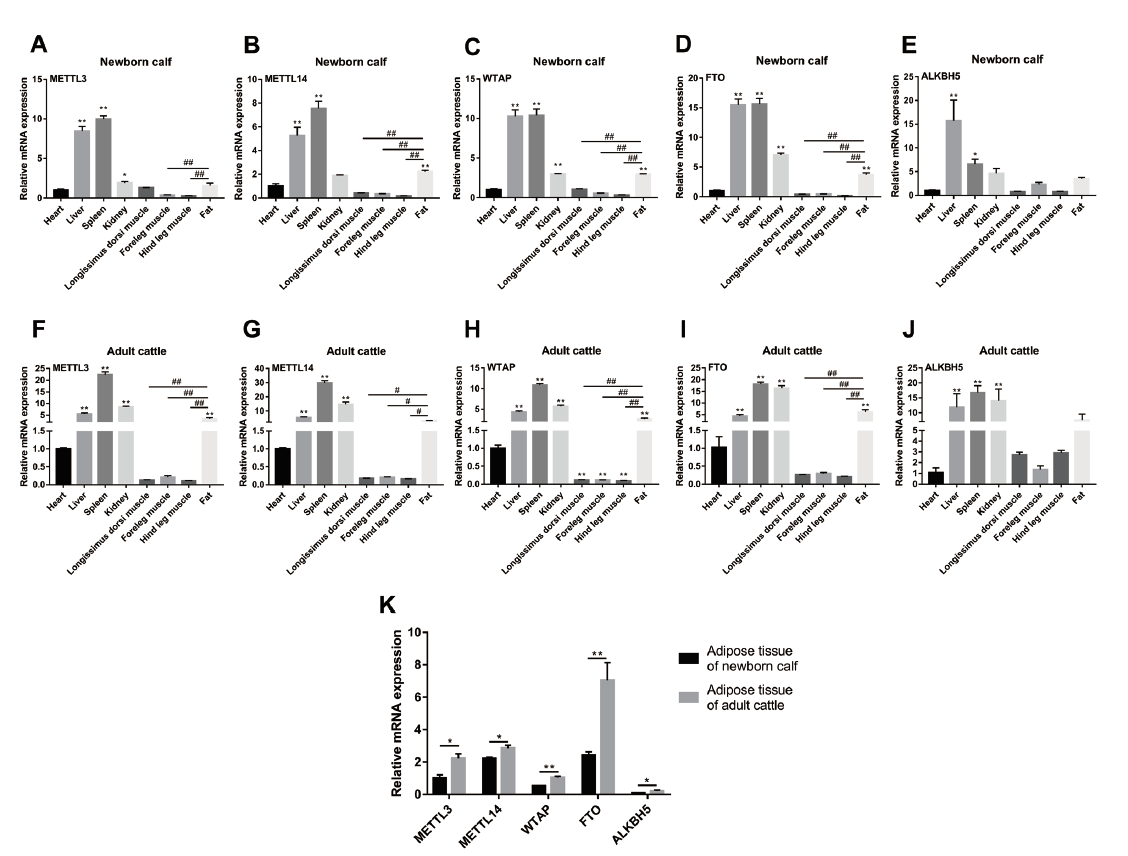
Fig. 1 Expressions of m6A methylation-related genes in different tissues of Qinchuan beef cattle A-E:Tissue mRNA expression profiles of METTL3(A),METTL14(B),WTAP(C),FTO(D)and ALKBH5(E)in newborn Qinchuan beef cattle. F-J:Tissue mRNA expression profiles of METTL3(F),METTL14(G),WTAP(H),FTO(I)and ALKBH5(J)in adult Qinchuan beef cattle. K:Comparison of expression levels of METTL3,METTL14,WTAP,FTO and ALKBH5 in the adipose tissues of newborn and adult Qinchuan beef cattle. A-J:*P < 0.05,** P < 0.01,other tissues versus heart;# P < 0.05,##P < 0.01,fat versus skeletal muscles;no superscripts indicate no significant difference(P > 0.05). K:*P < 0.05,** P < 0.01
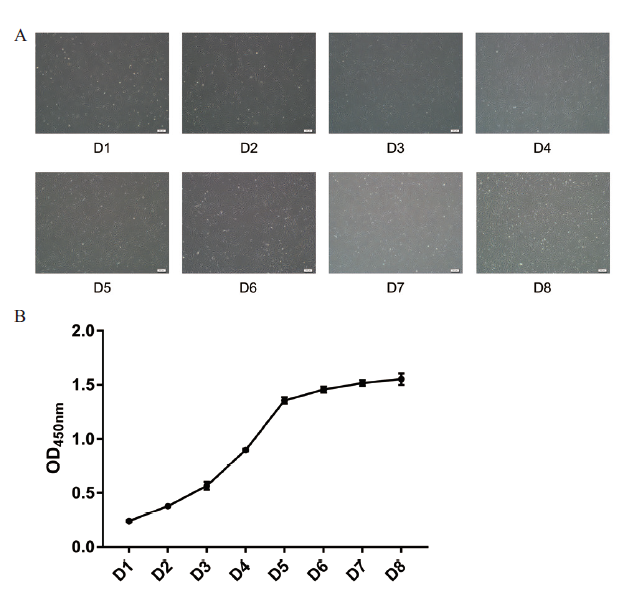
Fig. 2 Morphology and growth curve of preadipocytes from Qinchuan beef cattle in vitroA:Phenotypic observation of preadipocytes proliferation in Qinchuan beef cattle at different growth points(scale bar:200 μm). B:Growth curve of preadipocytes in Qinchuan beef cattle
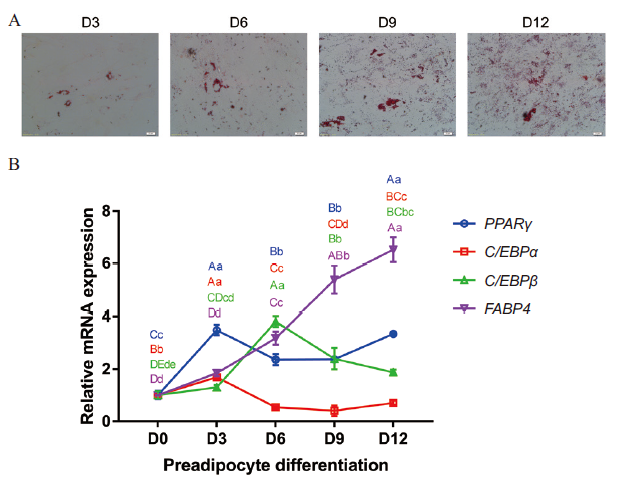
Fig. 3 Detection of preadipocytes adipogenic differentiation in Qinchuan beef cattle A:Observation of lipid droplet formation on day 3,6,9 and 12 of culture in differentiation medium after the induction of adipogenic differentiation(scale bar:20 μm);B:Relative mRNA expression of adipogenesis-related genes(PPARγ,C/EBPα,C/EBPβ and FABP4)during bovine preadipocytes differentiation. Different capital letters indicate very significant differences(P < 0.01),different lowercase letters indicate significant differences(P < 0.05),and the same letters indicate no significant differences(P > 0.05). The same below
| [1] | 昝林森, 王洪程, 梅楚刚. 秦川牛肉用选育改良及产业化开发[J]. 农业生物技术学报, 2015, 23(1):135-140. |
| Zan LS, Wang HC, Mei CG. Breeding and improvement of Qinchuan cattle and its beef industrialization[J]. J Agric Biotechnol, 2015, 23(1):135-140. | |
| [2] |
Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out[J]. Nat Rev Mol Cell Biol, 2006, 7(12):885-896.
doi: 10.1038/nrm2066 URL |
| [3] |
Farmer SR. Transcriptional control of adipocyte formation[J]. Cell Metab, 2006, 4(4):263-273.
doi: 10.1016/j.cmet.2006.07.001 URL |
| [4] |
Zhu JG, Xia L, Ji CB, et al. Differential DNA methylation status between human preadipocytes and mature adipocytes[J]. Cell Biochem Biophys, 2012, 63(1):1-15.
doi: 10.1007/s12013-012-9336-3 URL |
| [5] |
Wang LF, Jin QH, Lee JE, et al. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis[J]. PNAS, 2010, 107(16):7317-7322.
doi: 10.1073/pnas.1000031107 URL |
| [6] |
Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation[J]. Nat Chem Biol, 2014, 10(2):93-95.
doi: 10.1038/nchembio.1432 URL |
| [7] | Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase[J]. Cell Res, 2014, 24(2):177-189. |
| [8] |
Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO[J]. Nat Chem Biol, 2011, 7(12):885-887.
doi: 10.1038/nchembio.687 URL |
| [9] |
Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility[J]. Mol Cell, 2013, 49(1):18-29.
doi: 10.1016/j.molcel.2012.10.015 URL |
| [10] |
Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability[J]. Nature, 2014, 505(7481):117-120.
doi: 10.1038/nature12730 URL |
| [11] |
Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation[J]. Nat Rev Genet, 2014, 15(5):293-306.
doi: 10.1038/nrg3724 URL |
| [12] |
Wang X, Zhao BS, Roundtree IA, et al. N6-methyladenosine modulates messenger RNA translation efficiency[J]. Cell, 2015, 161(6):1388-1399.
doi: 10.1016/j.cell.2015.05.014 URL |
| [13] |
Shi H, Wei J, He C. Where, when, and how:context-dependent functions of RNA methylation writers, readers, and erasers[J]. Mol Cell, 2019, 74(4):640-650.
doi: 10.1016/j.molcel.2019.04.025 URL |
| [14] |
Frayling TM, Timpson NJ, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity[J]. Science, 2007, 316(5826):889-894.
doi: 10.1126/science.1141634 pmid: 17434869 |
| [15] |
Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis[J]. Cell Res, 2014, 24(12):1403-1419.
doi: 10.1038/cr.2014.151 pmid: 25412662 |
| [16] |
Wang X, Wu R, Liu Y, et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7[J]. Autophagy, 2020, 16(7):1221-1235.
doi: 10.1080/15548627.2019.1659617 URL |
| [17] |
Merkestein M, Laber S, McMurray F, et al. FTO influences adipogenesis by regulating mitotic clonal expansion[J]. Nat Commun, 2015, 6:6792.
doi: 10.1038/ncomms7792 pmid: 25881961 |
| [18] |
Kobayashi M, Ohsugi M, Sasako T, et al. The RNA methyltransferase complex of WTAP, METTL3, and METTL14 regulates mitotic clonal expansion in adipogenesis[J]. Mol Cell Biol, 2018, 38(16):e00116-18. DOI: 10.1128/mcb.00116-18.
doi: 10.1128/mcb.00116-18 |
| [19] |
Xie W, Ma LL, Xu YQ, et al. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism[J]. Biochem Biophys Res Commun, 2019, 518(1):120-126.
doi: 10.1016/j.bbrc.2019.08.018 URL |
| [20] | Yao Y, Bi Z, et al. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2-dependent manner[J]. FASEB J, 2019(6):7529-7544. |
| [21] |
Wang Y, Gao M, Zhu F, et al. METTL3 is essential for postnatal development of brown adipose tissue and energy expenditure in mice[J]. Nat Commun, 2020, 11(1):1648.
doi: 10.1038/s41467-020-15488-2 URL |
| [22] |
Tao XL, Chen JN, Jiang YZ, et al. Transcriptome-wide N ^ 6-methyladenosine methylome profiling of porcine muscle and adipose tissues reveals a potential mechanism for transcriptional regulation and differential methylation pattern[J]. BMC Genom, 2017, 18(1):1-13.
doi: 10.1186/s12864-016-3406-7 URL |
| [23] |
Cheng BH, Leng L, et al. Profiling of RNA N6-methyladenosine methylation reveals the critical role of m6A in chicken adipose deposition[J]. Front Cell Dev Biol, 2021, 9:590468.
doi: 10.3389/fcell.2021.590468 URL |
| [24] |
Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5' Terminus of HeLa cell messenger RNA[J]. Cell, 1975, 4(4):379-386.
pmid: 164293 |
| [25] |
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq[J]. Nature, 2012, 485(7397):201-206.
doi: 10.1038/nature11112 URL |
| [26] |
Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons[J]. Cell, 2012, 149(7):1635-1646.
doi: 10.1016/j.cell.2012.05.003 pmid: 22608085 |
| [27] |
Lin S, Choe J, Du P, et al. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells[J]. Mol Cell, 2016, 62(3):335-345.
doi: 10.1016/j.molcel.2016.03.021 URL |
| [28] |
Yang D, Qiao J, Wang G, et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential[J]. Nucleic Acids Res, 2018, 46(8):3906-3920.
doi: 10.1093/nar/gky130 URL |
| [29] |
Bertero A, Brown S, Madrigal P, et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency[J]. Nature, 2018, 555(7695):256-259.
doi: 10.1038/nature25784 URL |
| [30] |
Batista PJ, Molinie B, Wang J, et al. M(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells[J]. Cell Stem Cell, 2014, 15(6):707-719.
doi: 10.1016/j.stem.2014.09.019 pmid: 25456834 |
| [31] |
Zhang XX, Yao YL, Han JH, et al. Longitudinal epitranscriptome profiling reveals the crucial role of N6-methyladenosine methylation in porcine prenatal skeletal muscle development[J]. J Genet Genom, 2020, 47(8):466-476.
doi: 10.1016/j.jgg.2020.07.003 URL |
| [32] | Wang X, Huang N, et al. FTO is required for myogenesis by positively regulating mTOR-PGC-1α pathway-mediated mitochondria biogenesis[J]. Cell Death Dis, 2017, 8(3):e2702. |
| [33] |
Song T, Yang Y, Wei H, et al. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation[J]. Nucleic Acids Res, 2019, 47(12):6130-6144.
doi: 10.1093/nar/gkz312 URL |
| [34] |
Xu T, Xu Z,Lu L,et al. Transcriptome-wide study revealed m6A reg-ulation of embryonic muscle development in Dingan goose(Anser cygnoides orientalis)[J]. BMC Genomics, 2021, 22(1):270.
doi: 10.1186/s12864-021-07556-8 URL |
| [35] | 朱琳娜. FTO、METTL3基因表达对猪脂肪细胞mRNA N6-甲基腺苷水平及脂肪沉积的影响研究[D]. 杭州: 浙江大学, 2014. |
| Zhu LN. Effect of FTO, METTL3 Gene expression on mRNA m6A methylation and lipid metabolism in porcine subcutaneous adipocytes[D]. Hangzhou: Zhejiang University, 2014. | |
| [36] |
Fredriksson R, Hägglund M, Olszewski PK, et al. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain[J]. Endocrinology, 2008, 149(5):2062-2071.
doi: 10.1210/en.2007-1457 pmid: 18218688 |
| [37] | 马子玉. METTL3对猪干细胞多能性的调控作用[D]. 杨凌: 西北农林科技大学, 2018. |
| Ma ZY. METTL3 regulates the pluripotency of porcine pluripotent stem cells[D]. Yangling: Northwest A & F University, 2018. | |
| [38] |
Bravard A, Lefai E, et al. FTO is increased in muscle during type 2 diabetes, and its overexpression in myotubes alters insulin signaling, enhances lipogenesis and ROS production, and induces mitochondrial dysfunction[J]. Diabetes, 2011, 60(1):258-268.
doi: 10.2337/db10-0281 pmid: 20943749 |
| [39] |
Chen JQ, Zhou XH, et al. FTO-dependent function of N6-methyladenosine is involved in the hepatoprotective effects of betaine on adolescent mice[J]. J Physiol Biochem, 2015, 71(3):405-413.
doi: 10.1007/s13105-015-0420-1 URL |
| [40] |
Aggarwal S, Pittenger MF. Human mesenchymal stem cells modulate allogeneic immune cell responses[J]. Blood, 2005, 105(4):1815-1822.
doi: 10.1182/blood-2004-04-1559 URL |
| [41] |
Rodriguez AM, Elabd C, Delteil F, et al. Adipocyte differentiation of multipotent cells established from human adipose tissue[J]. Biochem Biophys Res Commun, 2004, 315(2):255-263.
doi: 10.1016/j.bbrc.2004.01.053 URL |
| [42] |
Xia T, Wu X, Cao M, et al. The RNA m6A methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion[J]. Pathol Res Pract, 2019, 215(11):152666.
doi: 10.1016/j.prp.2019.152666 URL |
| [43] |
Han J, Wang JZ, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner[J]. Mol Cancer, 2019, 18(1):110.
doi: 10.1186/s12943-018-0931-9 URL |
| [44] |
Liu J, et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer[J]. Nat Cell Biol, 2018, 20(9):1074-1083.
doi: 10.1038/s41556-018-0174-4 URL |
| [45] | 徐晓东. N6-甲基嘌呤(m6A)甲基转移酶METTL14对胰腺癌增殖和侵袭转移的影响及其机制研究[D]. 武汉: 华中科技大学, 2017. |
| Xu XD. The effect METTL14 on prolifetation and metastasis of pancreatic cancer cells and its mechanisms[D]. Wuhan: Huazhong University of Science and Technology, 2017. | |
| [46] |
Small TW, Bolender Z, Bueno C, et al. Wilms’ tumor 1-associating protein regulates the proliferation of vascular smooth muscle cells[J]. Circ Res, 2006, 99(12):1338-1346.
doi: 10.1161/01.RES.0000252289.79841.d3 URL |
| [47] |
Horiuchi K, Umetani M, Minami T, et al. Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA[J]. PNAS, 2006, 103(46):17278-17283.
doi: 10.1073/pnas.0608357103 pmid: 17088532 |
| [48] | Kong Y, Wu R, et al. Wilms’ tumor 1-associating protein contributes to psoriasis by promoting keratinocytes proliferation via regulating cyclinA2 and CDK2[J]. Int Immun, 2020, 88:106918. |
| [49] |
Su R, Dong L, Li C, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6a/MYC/CEBPA signaling[J]. Cell, 2018, 172(1/2):90-105. e23.
doi: 10.1016/j.cell.2017.11.031 URL |
| [50] |
Zhang S, Zhao BS, Zhou A, et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program[J]. Cancer Cell, 2017, 31(4):591-606. e6.
doi: 10.1016/j.ccell.2017.02.013 URL |
| [51] | 方婷晓. m6A去甲基化酶ALKBH5抑制食管鳞癌的增殖、侵袭和迁移[D]. 广州: 南方医科大学, 2019. |
| Fang TX. M6A demethylase ALKBH5 inhibits proliferation, migration and invasion of esophageal squamous cell carcinoma[D]. Guangzhou: Southern Medical University, 2019. | |
| [52] | Wu R, Liu Y, Yao Y, et al. FTO regulates adipogenesis by controlling cell cycle progression via m6A-YTHDF2 dependent mechanism[J]. Biochim Biophys Acta Mol Cell Biol Lipids, 2018, 1863(10):1323-1330. |
| [53] | 任倩. FTO通过抑制线粒体UPR缓解小鼠脂肪细胞凋亡的作用机制[D]. 杨凌: 西北农林科技大学, 2019. |
| Ren Q. The mechanism of FTO on suppressing mice adipocytes apoptosis by reducing mitochondrial UPR[D]. Yangling: Northwest A & F University, 2019. | |
| [54] |
Kang H, Zhang Z, Yu L, et al. FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation[J]. J Cell Biochem, 2018, 119(7):5676-5685.
doi: 10.1002/jcb.26746 URL |
| [55] |
Jia X, Nie Q, Lamont SJ, et al. Variation in sequence and expression of the avian FTO, and association with glucose metabolism, body weight, fatness and body composition in chickens[J]. Int J Obes:Lond, 2012, 36(8):1054-1061.
doi: 10.1038/ijo.2011.221 URL |
| [56] |
Cai M, Liu Q, Jiang Q, et al. Loss of m6 A on FAM134B promotes adipogenesis in porcine adipocytes through m6 A-YTHDF2-dependent way[J]. IUBMB Life, 2019, 71(5):580-586.
doi: 10.1002/iub.1974 URL |
| [57] |
Jiang Q, Sun B, Liu Q, et al. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m6A-YTHDF1-dependent mechanism[J]. FASEB J, 2019, 33(2):2971-2981.
doi: 10.1096/fj.201801393RRR pmid: 30339471 |
| [58] | 李冬. METTL3对奶牛乳腺上皮细胞乳蛋白和乳脂肪合成的调控及其作用机理[D]. 哈尔滨: 东北农业大学, 2017. |
| Li D. The regulatory mechanism of METTL3 on milk protein and milk fat synthesis in bovine mammary epithelial cells[D]. Harbin: Northeast Agricultural University, 2017. | |
| [59] |
Zhong X, Yu J, Frazier K, et al. Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation[J]. Cell Rep, 2018, 25(7):1816-1828. e4.
doi: 10.1016/j.celrep.2018.10.068 URL |
| [60] |
Tews D, Fischer-Posovszky P, Wabitsch M. Regulation of FTO and FTM expression during human preadipocyte differentiation[J]. Horm Metab Res, 2011, 43(1):17-21.
doi: 10.1055/s-0030-1265130 pmid: 20865646 |
| [61] | Friebe D, Löffler D, Schönberg M, et al. Impact of metabolic regulators on the expression of the obesity associated genes FTO and NAMPT in human preadipocytes and adipocytes[J]. PLoS One, 2011, 6(6):e19526. |
| [1] | ZHANG Yan-feng, DING Yan-ling, MA Ying, ZHOU Xiao-nan, YANG Chao-yun, SHI Yuan-gang, KANG Xiao-long. Comparative Analysis of Rumen and Fecal Microbial Characteristics Associated with Residual Feed Intake in Beef Cattle [J]. Biotechnology Bulletin, 2023, 39(1): 295-304. |
| [2] | XU Xiang, DONG Wei-peng, ZHANG Shao-hua, FENG Chen-yi, LIU Tian-fu, YAN Jiong. Construction of Fsp27 Gene Silencing Vector and Its Effect on Cell Lipolysis [J]. Biotechnology Bulletin, 2020, 36(1): 88-94. |
| [3] | ZHAO Rui-xiao, HAN Fei, JIANG Ming-feng, ZENG Lian, HAN Jia-yu, HUANG Ying, ZHOU Hang. Application of PCR-membrane Chip Technology in the Identification of Yak and Yak-cattle Meat [J]. Biotechnology Bulletin, 2020, 36(1): 202-208. |
| [4] | YANG Lei, YE Zhou-jie, LI Zhao-long, SHEN Yang-kun, FU Ya-juan. Effects of TET2 on T Cell Proliferation by Electroporation [J]. Biotechnology Bulletin, 2020, 36(1): 229-237. |
| [5] | HE Xiang-dong, XIA Yi, JIGE Mo-ti, WANG Hui, ZI Xiang-dong. mRNA Expressions of Hypoxia Inducible Factor Genes in Female Yak Reproductive System [J]. Biotechnology Bulletin, 2019, 35(4): 82-87. |
| [6] | SU Qiu-ju, ZHOU Xiang, LI Guang-peng, BAI Chun-ling, XU Wen-tao, LIU Bang. A New Method for Identifying and Genotyping MSTN Gene-edited Cattle [J]. Biotechnology Bulletin, 2019, 35(4): 208-212. |
| [7] | JI Wen-bo, WANG Hui, CHAI Zhi-xin, WANG Ji-kun, XIN Jin-wei, ZHONG Jin-cheng. Molecular Cloning and Tissue Expression of Gene HYOU1 [J]. Biotechnology Bulletin, 2019, 35(3): 123-131. |
| [8] | LI Ping ,ZHANG Gui-ping, HU Jian-ran. Effects of Total Flavonoids from Forsythia suspense on the Proliferation of Gastric Cancer Cell MGC80-3 [J]. Biotechnology Bulletin, 2018, 34(6): 199-203. |
| [9] | YANG Yuan-xiao, ZI Xiang-dong. Cloning of KISS-1 Gene in Yak and Its Expression in Reproductive Axis [J]. Biotechnology Bulletin, 2018, 34(3): 142-149. |
| [10] | GUO Hong-yan, GAO Han, WU Qi, SUN Xiao-jie, LIU Xiu-cai, ZHAO Li-qun. Construction of SGK3 Gene Lentiviral RNA Interference Vector and Effects on Cell proliferation and Apoptosis of Breast Cancer Cell Line MB-474 [J]. Biotechnology Bulletin, 2018, 34(1): 247-252. |
| [11] | HU Jian-ran LI Ping LEI Hai-ying LIU Xian-rui. Effects of Polysaccharides from Lu Dangshen(Codonopsis pilosula)on Proliferation and Migration of Human Cervical SiHa [J]. Biotechnology Bulletin, 2017, 33(5): 159-163. |
| [12] | Li Jiyou, Liang Chunhua, Liu Xia, Sun Xuejing, Du Xiaohua. Polymorphism Analysis of MSTN Gene Exons of Ganzhou Simmental Cattle in Gansu Province [J]. Biotechnology Bulletin, 2015, 31(6): 165-169. |
| [13] | Liang Chunhua, Liu Xia, Sun Xuejing, Luo Yuzhu, Gao Xuejin, Du Xiaohua. Genetic Variation Study of NGB Gene Exon 3 of Simmental Cattle Hybrid Taxon in Gansu Hexi Regions [J]. Biotechnology Bulletin, 2015, 31(2): 122-128. |
| [14] | Liu Fengxia, Liu Wenjuan, Li Jianyong, Chen Shenguo. A Study on the Regulation Mechanism of ERK1/2 Promoting Proliferation and Inhibiting Apoptosis in Esophageal Squamous Carcinoma [J]. Biotechnology Bulletin, 2015, 31(10): 242-248. |
| [15] | He Dongyang,Ma Chao,Gao Zhenyue,Wang Shuzhen,Chen Yijun. The Comparison of Expression Efficiency of Human CD137L in Different Expression System [J]. Biotechnology Bulletin, 2014, 0(9): 178-184. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||