








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (3): 109-117.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0681
Previous Articles Next Articles
ZHAO Hong-yuan1( ), LIU Qiang2(
), LIU Qiang2( ), CHENG Wen-yu1(
), CHENG Wen-yu1( )
)
Received:2023-07-17
Online:2024-03-26
Published:2024-04-08
Contact:
LIU Qiang, CHENG Wen-yu
E-mail:zhaohongyuan18@163.com;liuqiang@aku.edu.cn;wenyucheng1989@163.com
ZHAO Hong-yuan, LIU Qiang, CHENG Wen-yu. Research Progress in cGAS-STING Signaling Pathway in ASFV Antagonizing Host[J]. Biotechnology Bulletin, 2024, 40(3): 109-117.
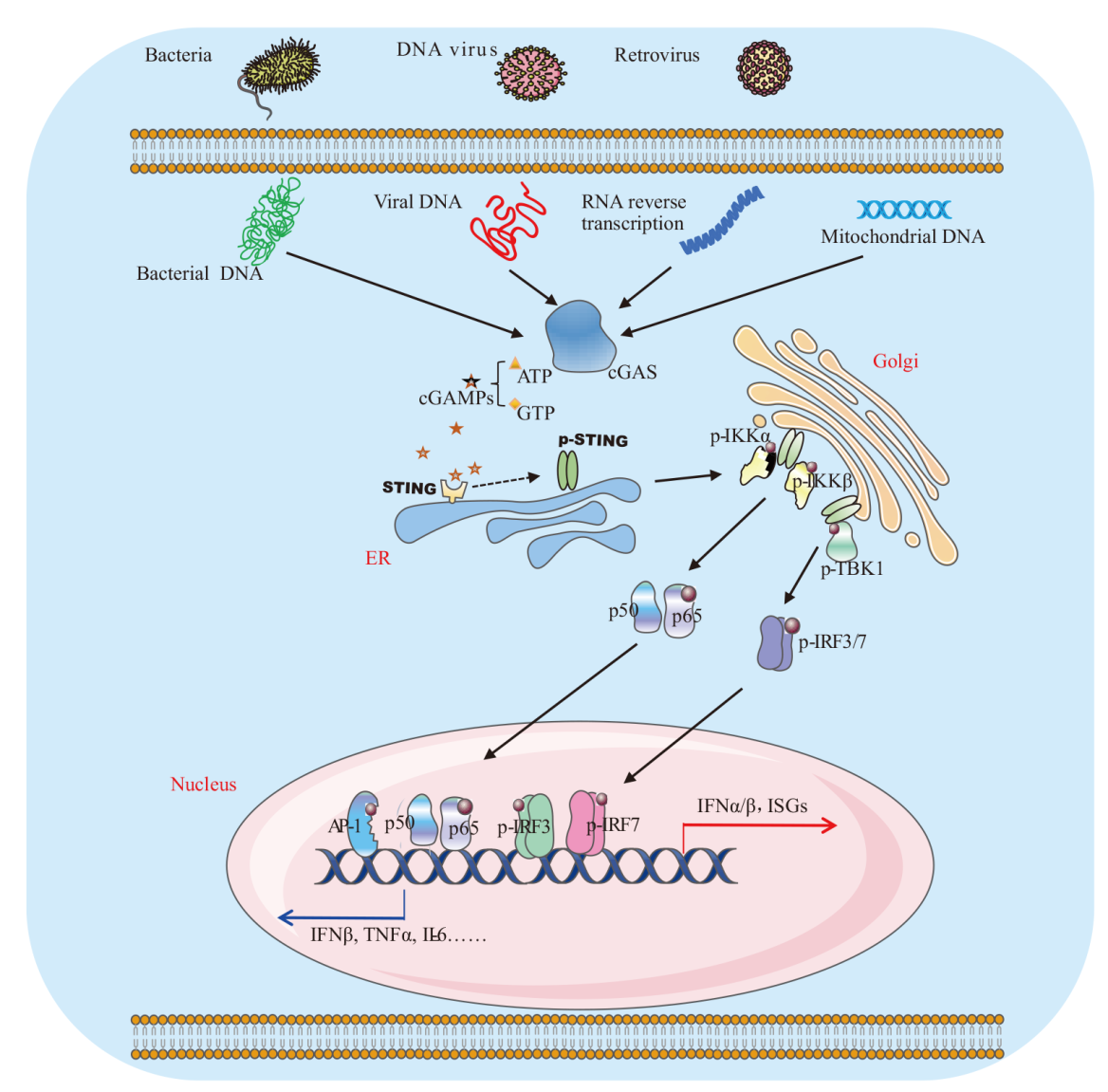
Fig. 1 Schematic diagram of cGAS-STING signal pathway AP-1: Activating protein 1. cGAS: Cyclic GMP-AMP synthase. cGAMP: Cyclic guanosine monophosphate-adenosine monophosphate. ER: Endoplasmic reticulum. IKK: Inhibitor of nuclear factor-κB kinase. IL: Interleukin. IRF: IFN-regulatory factor. ISG: Interferon stimulated gene. STING: Stimulator of interferon gene. p-STING: Phosphorylated STING. TBK1: TANK-binding kinase 1
| [1] |
赵鸿远, 王朝, 成温玉, 等. 抗非洲猪瘟病毒制剂的研究进展[J]. 生物技术通报, 2021, 37(5): 174-181.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-1079 |
| Zhao HY, Wang Z, Cheng WY, et al. Progress on antiviral agents against African swine fever virus[J]. Biotechnol Bull, 2021, 37(5): 174-181. | |
| [2] | Alejo A, Matamoros T, Guerra M, et al. A proteomic atlas of the African swine fever virus particle[J]. J Virol, 2018, 92(23): e01293-18. |
| [3] |
Cheng WY, He XB, Jia HJ, et al. The cGas-sting signaling pathway is required for the innate immune response against ectromelia virus[J]. Front Immunol, 2018, 9: 1297.
doi: 10.3389/fimmu.2018.01297 URL |
| [4] | 成温玉, 雷霆宇, 张博昕, 等. DNA识别受体介导的天然免疫参与抗痘病毒感染的研究进展[J]. 中国人兽共患病学报, 2022, 38(4): 341-348. |
| Cheng WY, Lei TY, Zhang BX, et al. Research progress in DNA sensor-mediated innate immunity against poxvirus infection[J]. Chin J Zoonoses, 2022, 38(4): 341-348. | |
| [5] | 成温玉, 何小兵, 贾怀杰, 等. 小鼠DDX5分子结构特征及功能初步探究[J]. 基因组学与应用生物学, 2021, 40(S1): 1983-1992. |
| Cheng WY, He XB, Jia HJ, et al. Analysis of molecular structural characteristics and elementary functions of mouse DDX5[J]. Genom Appl Biol, 2021, 40(S1): 1983-1992. | |
| [6] |
He WR, Yuan J, Ma YH, et al. Modulation of host antiviral innate immunity by African swine fever virus: a review[J]. Animals, 2022, 12(21): 2935.
doi: 10.3390/ani12212935 URL |
| [7] | Cheng ZL, Dai T, He XL, et al. The interactions between cGAS-STING pathway and pathogens[J]. Signal Transduct Target Ther, 2020, 5(1): 91. |
| [8] |
Andreeva L, Hiller B, Kostrewa D, et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders[J]. Nature, 2017, 549(7672): 394-398.
doi: 10.1038/nature23890 URL |
| [9] |
Luecke S, Holleufer A, Christensen MH, et al. cGAS is activated by DNA in a length-dependent manner[J]. EMBO Rep, 2017, 18(10): 1707-1715.
doi: 10.15252/embr.201744017 pmid: 28801534 |
| [10] |
Shang GJ, Zhang CG, Chen ZJ, et al. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP[J]. Nature, 2019, 567(7748): 389-393.
doi: 10.1038/s41586-019-0998-5 |
| [11] |
Hussain B, Xie YF, Jabeen U, et al. Activation of STING based on its structural features[J]. Front Immunol, 2022, 13: 808607.
doi: 10.3389/fimmu.2022.808607 URL |
| [12] |
Balka KR, Louis C, Saunders TL, et al. TBK1 and IKKε act redundantly to mediate STING-induced NF-κB responses in myeloid cells[J]. Cell Rep, 2020, 31(1): 107492.
doi: 10.1016/j.celrep.2020.03.056 URL |
| [13] | García-Belmonte R, Pérez-Núñez D, Pittau M, et al. African swine fever virus Armenia/07 virulent strain controls interferon beta production through the cGAS-STING pathway[J]. J Virol, 2019, 93(12): e02298-18. |
| [14] |
Karalyan Z, Zakaryan H, Sargsyan K, et al. Interferon status and white blood cells during infection with African swine fever virus in vivo[J]. Vet Immunol Immunopathol, 2012, 145(1-2): 551-555.
doi: 10.1016/j.vetimm.2011.12.013 URL |
| [15] |
Franzoni G, Pedrera M, Sánchez-Cordón PJ. African swine fever virus infection and cytokine response in vivo: an update[J]. Viruses, 2023, 15(1): 233.
doi: 10.3390/v15010233 URL |
| [16] |
Eaglesham JB, Pan YD, Kupper TS, et al. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling[J]. Nature, 2019, 566(7743): 259-263.
doi: 10.1038/s41586-019-0928-6 |
| [17] |
Dodantenna N, Ranathunga L, Chathuranga WAG, et al. African swine fever virus EP364R and C129R target cyclic GMP-AMP to inhibit the cGAS-STING signaling pathway[J]. J Virol, 2022, 96(15): e0102222.
doi: 10.1128/jvi.01022-22 URL |
| [18] |
Zheng WL, Xia NW, Zhang JJ, et al. African swine fever virus structural protein p17 inhibits cGAS-STING signaling pathway through interacting with STING[J]. Front Immunol, 2022, 13: 941579.
doi: 10.3389/fimmu.2022.941579 URL |
| [19] |
Zhu ZX, Li SS, Ma CN, et al. African swine fever virus E184L protein interacts with innate immune adaptor STING to block IFN production for viral replication and pathogenesis[J]. J Immunol, 2023, 210(4): 442-458.
doi: 10.4049/jimmunol.2200357 pmid: 36602826 |
| [20] |
Ramirez-Medina E, Vuono E, Rai A, et al. Deletion of E184L, a putative DIVA target from the pandemic strain of African swine fever virus, produces a reduction in virulence and protection against virulent challenge[J]. J Virol, 2022, 96(1): e0141921.
doi: 10.1128/JVI.01419-21 URL |
| [21] | 王曼, 沈宇清. 非洲猪瘟病毒结构蛋白CD2v的功能研究进展[J]. 中国免疫学杂志, 2021, 37(22): 2734-2737, 2744. |
| Wang M, Shen YQ. Research progress in function of ASFV structural protein CD2v[J]. Chin J Immunol, 2021, 37(22): 2734-2737, 2744. | |
| [22] |
Huang L, Chen WY, Liu HY, et al. African swine fever virus HLJ/18 CD2v suppresses type I IFN production and IFN-stimulated genes expression through negatively regulating cGMP-AMP synthase-STING and IFN signaling pathways[J]. J Immunol, 2023, 210(9): 1338-1350.
doi: 10.4049/jimmunol.2200813 pmid: 36971697 |
| [23] |
Li D, Yang WP, Li LL, et al. African swine fever virus MGF-505-7R negatively regulates cGAS-STING-mediated signaling pathway[J]. J Immunol, 2021, 206(8): 1844-1857.
doi: 10.4049/jimmunol.2001110 pmid: 33712518 |
| [24] |
Yang KD, Huang QT, Wang RY, et al. African swine fever virus MGF505-11R inhibits type I interferon production by negatively regulating the cGAS-STING-mediated signaling pathway[J]. Vet Microbiol, 2021, 263: 109265.
doi: 10.1016/j.vetmic.2021.109265 URL |
| [25] |
Chapman DAG, Tcherepanov V, Upton C, et al. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates[J]. J Gen Virol, 2008, 89(Pt 2): 397-408.
doi: 10.1099/vir.0.83343-0 pmid: 18198370 |
| [26] |
Cheng MY, Kanyema MM, Sun Y, et al. African swine fever virus L83L negatively regulates the cGAS-STING-mediated IFN-I pathway by recruiting tollip to promote STING autophagic degradation[J]. J Virol, 2023, 97(2): e0192322.
doi: 10.1128/jvi.01923-22 URL |
| [27] | Li JN, Song J, Kang L, et al. pMGF505-7 R determines pathogenicity of African swine fever virus infection by inhibiting IL-1β and type I IFN production[J]. PLoS Pathog, 2021, 17(7): e1009733. |
| [28] | O'Donnell V, Risatti GR, Holinka LG, et al. Simultaneous deletion of the 9GL and UK genes from the African swine fever virus Georgia 2007 isolate offers increased safety and protection against homologous challenge[J]. J Virol, 2016, 91(1): e01760-16. |
| [29] |
Wang XX, Wu J, Wu YT, et al. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1[J]. Biochem Biophys Res Commun, 2018, 506(3): 437-443.
doi: 10.1016/j.bbrc.2018.10.103 URL |
| [30] |
Huang L, Xu WJ, Liu HY, et al. African swine fever virus pI215L negatively regulates cGAS-STING signaling pathway through recruiting RNF138 to inhibit K63-linked ubiquitination of TBK1[J]. J Immunol, 2021, 207(11): 2754-2769.
doi: 10.4049/jimmunol.2100320 pmid: 34759016 |
| [31] |
Freitas FB, Frouco G, Martins C, et al. African swine fever virus encodes for an E2-ubiquitin conjugating enzyme that is mono- and di-ubiquitinated and required for viral replication cycle[J]. Sci Rep, 2018, 8(1): 3471.
doi: 10.1038/s41598-018-21872-2 pmid: 29472632 |
| [32] |
Sun MW, Yu SX, Ge HL, et al. The A137R protein of African swine fever virus inhibits type I interferon production via the autophagy-mediated lysosomal degradation of TBK1[J]. J Virol, 2022, 96(9): e0195721.
doi: 10.1128/jvi.01957-21 URL |
| [33] |
Reis AL, Abrams CC, Goatley LC, et al. Deletion of African swine fever virus interferon inhibitors from the genome of a virulent isolate reduces virulence in domestic pigs and induces a protective response[J]. Vaccine, 2016, 34(39): 4698-4705.
doi: S0264-410X(16)30668-5 pmid: 27521231 |
| [34] | Zhang KS, Yang B, Shen CC, et al. MGF360-9 L is a major virulence factor associated with the African swine fever virus by antagonizing the JAK/STAT signaling pathway[J]. mBio, 2022, 13(1): e0233021. |
| [35] |
Yang KD, Xue Y, Niu H, et al. African swine fever virus MGF360-11L negatively regulates cGAS-STING-mediated inhibition of type I interferon production[J]. Vet Res, 2022, 53(1): 7.
doi: 10.1186/s13567-022-01025-0 pmid: 35073979 |
| [36] |
Zhuo YS, Guo ZH, Ba TT, et al. African swine fever virus MGF360-12L inhibits type I interferon production by blocking the interaction of importin α and NF-κB signaling pathway[J]. Virol Sin, 2021, 36(2): 176-186.
doi: 10.1007/s12250-020-00304-4 pmid: 33141406 |
| [37] |
Wang Y, Cui S, Xin T, et al. African swine fever virus MGF360-14L negatively regulates type I interferon signaling by targeting IRF3[J]. Front Cell Infect Microbiol, 2022, 11: 818969.
doi: 10.3389/fcimb.2021.818969 URL |
| [38] |
Luo J, Zhang JJ, Ni JH, et al. The African swine fever virus protease pS273R inhibits DNA sensing cGAS-STING pathway by targeting IKKε[J]. Virulence, 2022, 13(1): 740-756.
doi: 10.1080/21505594.2022.2065962 URL |
| [39] | Li GB, Liu XX, Yang MY, et al. Crystal structure of African swine fever virus pS273R protease and implications for inhibitor design[J]. J Virol, 2020, 94(10): e02125-19. |
| [40] |
Zhao GH, Li TT, Liu XM, et al. African swine fever virus cysteine protease pS273R inhibits pyroptosis by noncanonically cleaving gasdermin D[J]. J Biol Chem, 2022, 298(1): 101480.
doi: 10.1016/j.jbc.2021.101480 URL |
| [41] |
Liu HS, Zhu ZX, Feng T, et al. African swine fever virus E120R protein inhibits interferon beta production by interacting with IRF3 to block its activation[J]. J Virol, 2021, 95(18): e0082421.
doi: 10.1128/JVI.00824-21 URL |
| [42] |
Zhang YY, Ke JN, Zhang JY, et al. African swine fever virus bearing an I226R gene deletion elicits robust immunity in pigs to African swine fever[J]. J Virol, 2021, 95(23): e0119921.
doi: 10.1128/JVI.01199-21 URL |
| [43] |
Hong JX, Chi XJ, Yuan X, et al. I226R protein of African swine fever virus is a suppressor of innate antiviral responses[J]. Viruses, 2022, 14(3): 575.
doi: 10.3390/v14030575 URL |
| [44] |
Cui S, Wang Y, Gao XT, et al. African swine fever virus M1249L protein antagonizes type I interferon production via suppressing phosphorylation of TBK1 and degrading IRF3[J]. Virus Res, 2022, 319: 198872.
doi: 10.1016/j.virusres.2022.198872 URL |
| [45] |
Liu XH, Liu HY, Ye GQ, et al. African swine fever virus pE301R negatively regulates cGAS-STING signaling pathway by inhibiting the nuclear translocation of IRF3[J]. Vet Microbiol, 2022, 274: 109556.
doi: 10.1016/j.vetmic.2022.109556 URL |
| [46] |
Chen H, Wang ZZ, Gao XY, et al. ASFV pD345L protein negatively regulates NF-κB signalling by inhibiting IKK kinase activity[J]. Vet Res, 2022, 53(1): 32.
doi: 10.1186/s13567-022-01050-z pmid: 35461299 |
| [47] |
Barrado-Gil L, Del Puerto A, Galindo I, et al. African swine fever virus ubiquitin-conjugating enzyme is an immunomodulator targeting NF-κB activation[J]. Viruses, 2021, 13(6): 1160.
doi: 10.3390/v13061160 URL |
| [1] | YE Hong, WANG Yu-kun. Research Progress in Immune Receptor Functions of Pattern-Recognition Receptor in Plants [J]. Biotechnology Bulletin, 2023, 39(12): 1-15. |
| [2] | CHEN Ying, WANG Yi-lei, ZOU Peng-fei. Cloning and Expression Analysis of TRAF6 from Large Yellow Croaker Larimichthys crocea [J]. Biotechnology Bulletin, 2022, 38(8): 233-243. |
| [3] | ZHAO Hong-yuan, WANG Zhao, CHENG Wen-yu, MA Ning-ning, LI Man, WEI Xiao-li. Progress on Antiviral Agents Against African Swine Fever Virus [J]. Biotechnology Bulletin, 2021, 37(5): 174-181. |
| [4] | ZOU Chen-chen, RUAN Ling-wei, SHI Hong. Wnt Signaling Pathway and Innate Immunity of Invertebrate [J]. Biotechnology Bulletin, 2021, 37(5): 182-196. |
| [5] | LI Kai-qing, LI Ying, WANG Yi-lei, ZOU Peng-fei. The Function of Receptor-interacting Protein(RIP)Kinases and the Research Progress in Teleost Fish [J]. Biotechnology Bulletin, 2021, 37(5): 197-211. |
| [6] | WANG Cai-xia, DU Fang-yuan, LIN Xiang-mei, Grzegorz Wozniakowski, WANG Qin, FENG Chun-yan, WU Shao-qiang. Generation of a Vero Cell Line Stably Expressing African Swine Fever Virus P54 Protein [J]. Biotechnology Bulletin, 2020, 36(5): 139-144. |
| [7] | HU Qi-chao, LUORENG Zhuo-ma, WEI Da-wei, YANG Jian, JIA Li, WANG Xing-ping, MA Yun. Research Progress on Innate Immunity-Related Coding Genes in the Regulation of Cow Mastitis [J]. Biotechnology Bulletin, 2020, 36(12): 239-246. |
| [8] | OU Yun-wen, LIU Li-jun, DAI Jun-fei, MA Bing, ZHANG Yong-guang, ZHANG Jie. Roles of African Swine Fever Virus Structural Proteins in Viral Infection [J]. Biotechnology Bulletin, 2019, 35(6): 156-163. |
| [9] | DONG Ru, CAO Yang-rong. Research Progress on the Immune Regulation of Symbiotic Nitrogen Fixation Between Legumes and Rhizobia [J]. Biotechnology Bulletin, 2019, 35(10): 25-33. |
| [10] | XIA Hong-li, CHENG Jun, YU Da-peng, CHEN Wen-jie, LU Yi-shan. Research Progress on Peptidoglycan Recognition Proteins in Fish [J]. Biotechnology Bulletin, 2018, 34(8): 58-66. |
| [11] | LUO Man, WANG Yu, HU Ya, XIU Jiang-fan, WANG Tao, PENG Jian, SHANG Xiao-li, WU Jian-wei. Cloning,Expression and Bacterial Binding of Peptidoglycan Recognition Proteins-SA Gene from Musca domestica [J]. Biotechnology Bulletin, 2016, 32(8): 145-151. |
| [12] | Bi Caihong, Zhang Qiuxia, Yu Shanshan, Chen Xuezhao, Liu Chunying, Zhu Qian. Molecular Cloning and Expression Analysis of MyD88 in Roughskin Soulpin,Trachidermus fasciatus [J]. Biotechnology Bulletin, 2015, 31(2): 135-142. |
| [13] | Xiu Jiangfan, Wei Chuanchuan, Chen Mingming, Wu Jianwei. Expression Analysis of Immune Genes at Different Induction Conditions of Third Instar Larvae of Musca domestica [J]. Biotechnology Bulletin, 2014, 0(6): 120-127. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||