








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 235-246.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0158
Previous Articles Next Articles
ZHOU Jiang-hong( ), XIA Fei, ZHONG Li, QIU Lan-fen, LI Guang, LIU Qian, ZHANG Guo-feng, SHAO Jin-li, LI Na, CHE Shao-chen(
), XIA Fei, ZHONG Li, QIU Lan-fen, LI Guang, LIU Qian, ZHANG Guo-feng, SHAO Jin-li, LI Na, CHE Shao-chen( )
)
Received:2024-02-18
Online:2024-07-26
Published:2024-07-30
Contact:
CHE Shao-chen
E-mail:zhoujh416@163.com;cheshaochen@163.com
ZHOU Jiang-hong, XIA Fei, ZHONG Li, QIU Lan-fen, LI Guang, LIU Qian, ZHANG Guo-feng, SHAO Jin-li, LI Na, CHE Shao-chen. Whole Genome Sequencing and Comparative Genomic Analysis of Antagonistic Bacterium CCBC3-3-1 against Verticillium dahlia[J]. Biotechnology Bulletin, 2024, 40(7): 235-246.
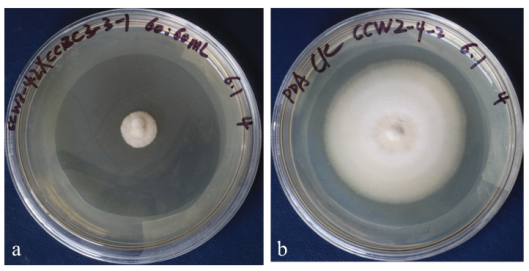
Fig. 1 Antagonism of the sterile fermentation broth of endogenous bacterium CCBC3-3-1 in Cotinus cog-gygria on V. dahlia a: The sterile fermentation broth of CCBC3-3-1 effectively inhibited the growth of V. dahlia. b: Control
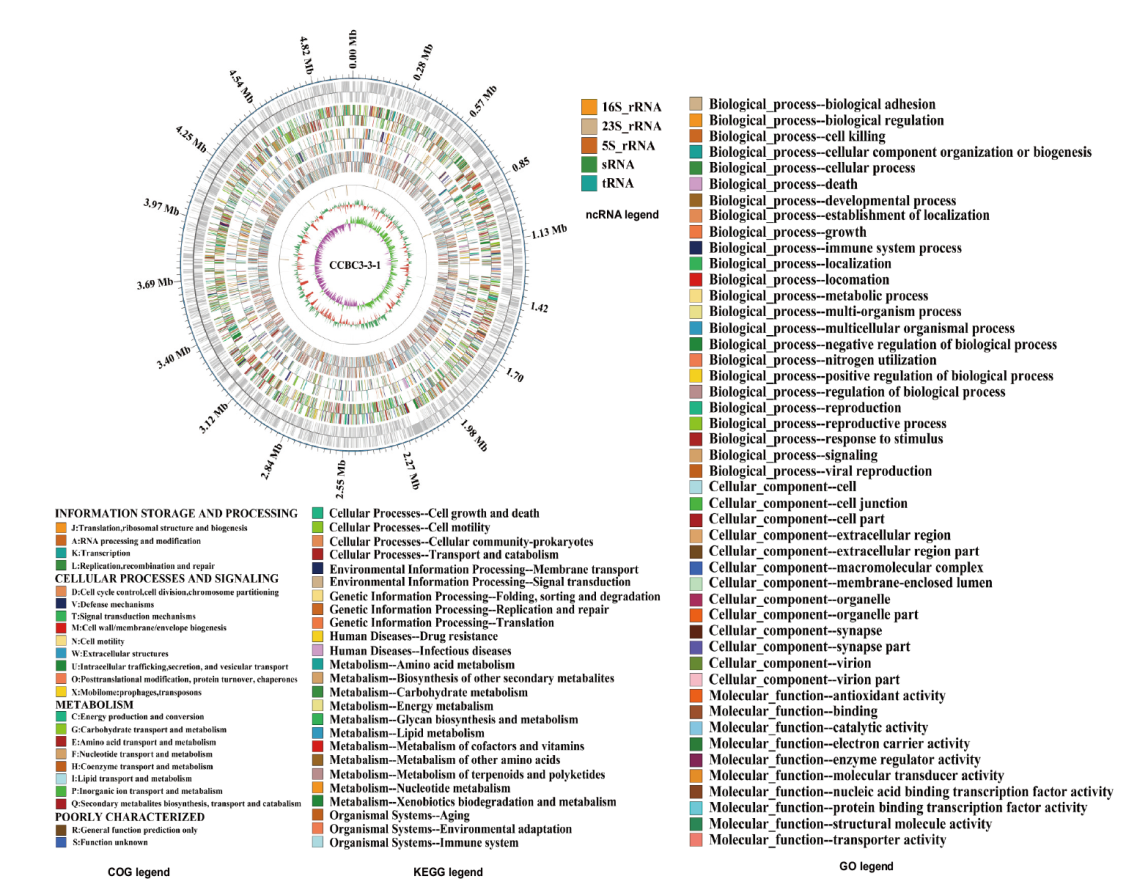
Fig. 2 Chromosome genome map of strain CCBC3-3-1 From outer to inner: 1: Genome size(million bp); 2: coding genes; 3: gene function annotation in the COG database; 4: gene function annotation in the KEGG database; 5: gene function annotation in the GO database; 6: nc RNA; 7: GC content(The inward red part indicates that the GC content in this region is lower than the average GC content of the entire genome, while the outward green part is the opposite. The higher the peak, the greater the difference between the GC content and the average GC content); 8: GC-skew(The algorithm is G-C/G+C, which is used to measure the relative content of G and C. If G>C, the GC-skew value is positive, represented by the inward pink part; if G<C, the GC-skew value is negative, represented by the outward light green part)

Fig. 3 Plasmid genome map of strain CCBC3-3-1 From outer to inner: 1: Gene function annotation in the COG database(Clockwise arrows indicate positive chain gene); 2: genome Size(kilo bp); 3: GC content(The inward red part indicates that the GC content in this region is lower than the average GC content of the entire genome, while the outward green part is the opposite. The higher the peak, the greater the difference between the GC content and the average GC content); 4: GC-skew(The algorithm is G-C/G+C, which is used to measure the relative content of G and C. If G>C, the GC-skew value is positive, represented by the inward pink part; if G<C, the GC-skew value is negative, represented by the outward light green part)
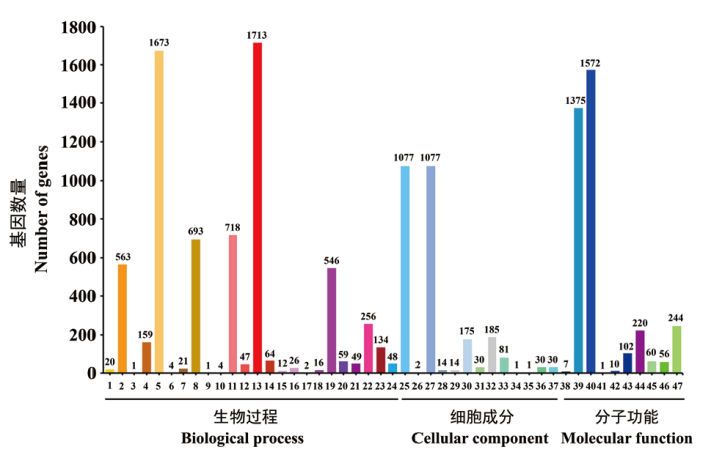
Fig. 5 GO function classification of strain CCBC3-3-1 1: Biological adhesion. 2: Biological regulation. 3: Cell killing. 4: Cellular component organization or biogenesis. 5: Cellular process. 6: Death. 7: Developmental process. 8: Establishment of localization. 9: Growth. 10: Immune system process. 11: Localization. 12: Locomotion. 13: Metabolic process. 14: Multi-organism process. 15: Multi cellular organismal process. 16: Negative regulation of biological process. 17: Nitrogen utilization. 18: Positive regulation of biological process. 19: Regulation of biological process. 20: Reproduction. 21: Reproductive process. 22: Response to stimulus. 23: Signaling. 24: Viral reproduction. 25: Cell. 26: Cell junction. 27: Cell part. 28: Extracellular region. 29: Extracellular region part. 30: Macromolecular complex. 31: Membrane-enclosed lumen. 32: Organelle. 33: Organelle part. 34: Synapse. 35: Synapse part. 36: Virion. 37: Virion part. 38: Antioxidant activity. 39: Binding. 40: Catalytic activity. 41: Electron carrier activity. 42: Enzyme regulator activity. 43: Molecular transducer activity. 44: Nucleic acid binding transcription factor activity. 45: Protein binding transcription factor activity. 46: Structural molecule activity. 47: Transporter activity
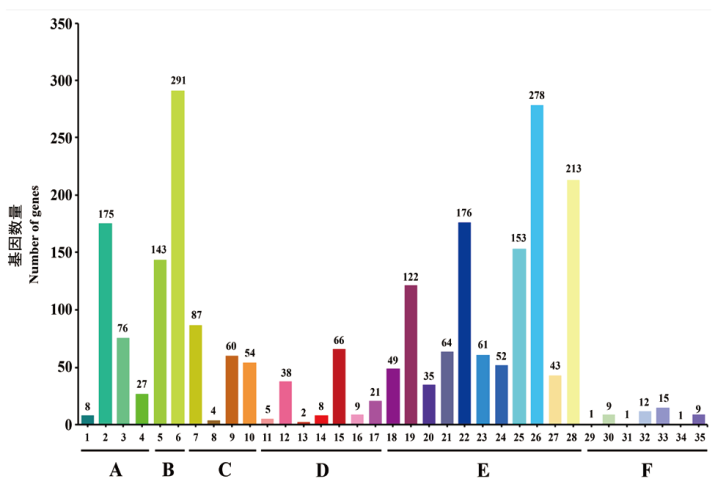
Fig. 6 KEEG function classification of strain CCBC3-3-1 A: Cellular processes(1: Transport and catabolism. 2: Cellular community-prokaryotes. 3: Cell motility. 4: Cell growth and death). B: Environmental information processing(5: Signal transduction. 6: Membrane transport). C: Genetic information processing(7: Translation. 8: Transcription. 9: Replication and repair. 10: Folding, sorting and degradation). D: Human diseases(11: Neurodegenerative diseases. 12: Infectious diseases. 13: Immune diseases. 14: Endocrine and metabolic diseases. 15: Drug resistance. 16: Cardiovascular diseases. 17: Cancers). E: Metabolism(18: Xenobiotics biodegradation and metabolism. 19: Nucleotide metabolism. 20: Metabolism of terpenoids and polyketides. 21: Metabolism of other amino acids. 22: Metabolism of cofactors and vitamins. 23: Lipid metabolism. 24: Glycan biosynthesis and metabolism. 25: Energy metabolism. 26: Carbohydrate metabolism. 27: Biosynthesis of other secondary metabolites. 28: Amino acid metabolism).F: Organismal systems(29: Nervous system. 30: Immune system. 31: Excretory system. 32: Environmental adaptation. 33: Endocrine system. 34: Digestive system. 35: Aging)
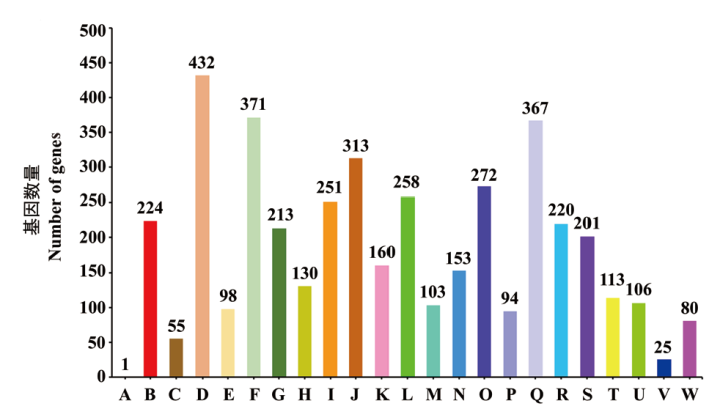
Fig. 7 COG function classification of strain CCBC3-3-1 A: RNA processing and modification. B: Energy production and conversion. C: Cell cycle control, cell division, chromosome partitioning. D: Amino acid transport and metabolism. E: Nucleotide transport and metabolism. F: Carbohydrate transport and metabolism. G: Coenzyme transport and metabolism. H: Lipid transport and metabolism. I: Translation, ribosomal structure and biogenesis. J: Transcription. K: Replication, recombination and repair. L: Cell wall/membrane/envelope biogenesis. M: Cell motility. N: Posttranslational modification, protein turnover, chaperones. O: Inorganic ion transport and metabolism. P: Secondary metabolites biosynthesis, transport and catabolism. Q: General function prediction only. R: Function unknown. S: Signal transduction mechanisms. T: Intracellular trafficking, secretion, and vesicular. U: Defense mechanisms. V: Extracellular structures. W: Mobilome: prophages, transposons

Fig. 8 CAZy function classification of strain CCBC3-3-1 AAs: Auxiliary activities. CBMs: Carbohydrate-binding modules. CEs: Carbohydrate esterases. GHs: Glycoside hydrolases.GTs: Glycosyl transferases. PLs: Polysaccharide lyases
| 项目Item | CCBC3-3-1 | P. vagans C9-1 | P. agglomerans TH81 | P. ananatis PA13 | P. eucalypti LMG24197 |
|---|---|---|---|---|---|
| GeneBank number | CP034363.1 | CP002206.1 | CP031649.1 | CP003085.1 | CP045720.1 |
| Length of chromosome /bp | 5 008 525 | 4 024 986 | 4 128 817 | 4 586 378 | 4 035 995 |
| GC content/% | 52.9 | 55.5 | 55.2 | 53.7 | 54.6 |
| Number of CDSs | 4 610 | 3 707 | 3 766 | 4 254 | 3 692 |
| Number of rRNAs | 22 | 22 | 21 | 22 | 21 |
| Number of tRNAs | 87 | 78 | 72 | 86 | 76 |
| Number of CRISPRS | 1 | 0 | 0 | 0 | 0 |
Table 1 Comparison of chromosome sequence feature between strain CCBC3-3-1 with other 4 species of Pantoea
| 项目Item | CCBC3-3-1 | P. vagans C9-1 | P. agglomerans TH81 | P. ananatis PA13 | P. eucalypti LMG24197 |
|---|---|---|---|---|---|
| GeneBank number | CP034363.1 | CP002206.1 | CP031649.1 | CP003085.1 | CP045720.1 |
| Length of chromosome /bp | 5 008 525 | 4 024 986 | 4 128 817 | 4 586 378 | 4 035 995 |
| GC content/% | 52.9 | 55.5 | 55.2 | 53.7 | 54.6 |
| Number of CDSs | 4 610 | 3 707 | 3 766 | 4 254 | 3 692 |
| Number of rRNAs | 22 | 22 | 21 | 22 | 21 |
| Number of tRNAs | 87 | 78 | 72 | 86 | 76 |
| Number of CRISPRS | 1 | 0 | 0 | 0 | 0 |
| 质粒 Plasmid | 数量 Number | GenBank登录号 GenBank number | 长度 Length/bp | GC含量 GC Content /% | CDSs数量 CDSs number | rRNA数量 rRNA number | tRNA数量 tRNA number | CRISPRS数量 CRISPRS number |
|---|---|---|---|---|---|---|---|---|
| CCBC3-3-1 | 3 | CP034364.1 CP034365.1 CP034366.1 | 25 471 29 467 96 304 | 46.7 42.7 49.9 | 38 37 114 | 0 0 0 | 0 0 0 | 0 0 0 |
| P. vagans C9-1 | 3 | CP001893.1 CP001894.1 CP001895.1 | 167 983 165 693 529 676 | 53.0 51.1 53.9 | 144 183 508 | 0 0 0 | 0 0 0 | 0 0 0 |
| P. agglomerans TH81 | 3 | CP031650.1 CP031651.1 CP031652.1 | 520 959 182 169 152 523 | 53.7 52.3 48.9 | 517 161 160 | 0 0 0 | 0 0 0 | 0 0 0 |
| P. ananatis PA13 | 1 | CP003086.1 | 280 753 | 52.3 | 257 | 0 | 0 | 0 |
| P. eucalypti LMG24197 | 3 | CP045721.1 CP045722.1 CP045723.1 | 529 303 138 725 94 967 | 52.2 50.9 52.2 | 528 114 99 | 0 0 0 | 0 0 0 | 0 0 0 |
Table 2 Comparison of strain CCBC3-3-1 plasmid sequence with other 4 species of Pantoea
| 质粒 Plasmid | 数量 Number | GenBank登录号 GenBank number | 长度 Length/bp | GC含量 GC Content /% | CDSs数量 CDSs number | rRNA数量 rRNA number | tRNA数量 tRNA number | CRISPRS数量 CRISPRS number |
|---|---|---|---|---|---|---|---|---|
| CCBC3-3-1 | 3 | CP034364.1 CP034365.1 CP034366.1 | 25 471 29 467 96 304 | 46.7 42.7 49.9 | 38 37 114 | 0 0 0 | 0 0 0 | 0 0 0 |
| P. vagans C9-1 | 3 | CP001893.1 CP001894.1 CP001895.1 | 167 983 165 693 529 676 | 53.0 51.1 53.9 | 144 183 508 | 0 0 0 | 0 0 0 | 0 0 0 |
| P. agglomerans TH81 | 3 | CP031650.1 CP031651.1 CP031652.1 | 520 959 182 169 152 523 | 53.7 52.3 48.9 | 517 161 160 | 0 0 0 | 0 0 0 | 0 0 0 |
| P. ananatis PA13 | 1 | CP003086.1 | 280 753 | 52.3 | 257 | 0 | 0 | 0 |
| P. eucalypti LMG24197 | 3 | CP045721.1 CP045722.1 CP045723.1 | 529 303 138 725 94 967 | 52.2 50.9 52.2 | 528 114 99 | 0 0 0 | 0 0 0 | 0 0 0 |
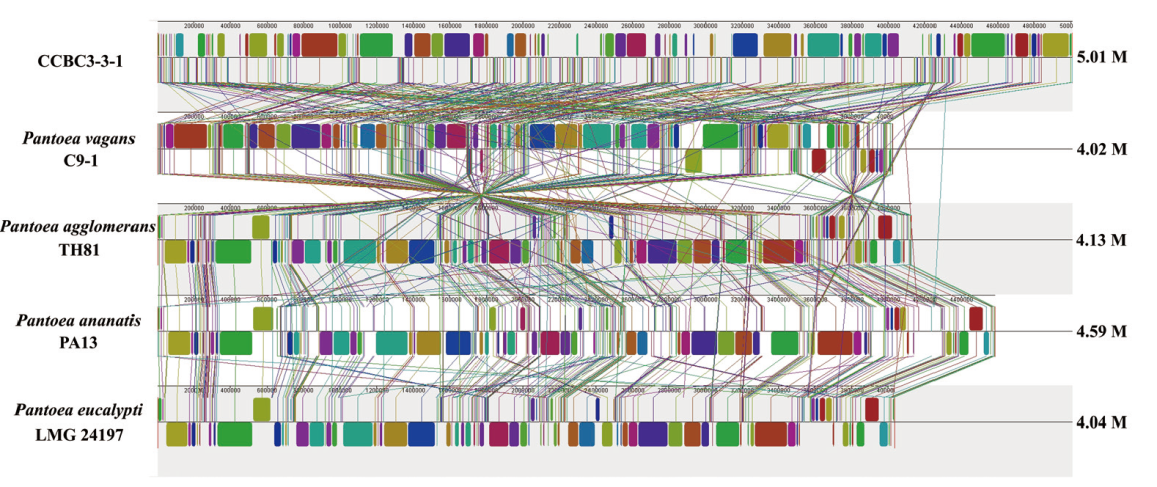
Fig. 9 Genomic collinearity analysis of CCBC3-3-1 with P. vagans C9-1, P. ananatis PA13, P. agglomerans TH81 and P. eu-calypti LMG24197 The same color modules joined by linear indicate collinear region
| 物种 Species | CCBC3-3-1 | |
|---|---|---|
| OrthoANIu/% | dDDH/% | |
| P. agglomerans TH81 | 75.95 | 20.90 |
| P. agglomerans C410P1 | 75.66 | 21.00 |
| P. agglomerans CHTF15 | 75.93 | 20.80 |
| P. agglomerans CQ10 | 76.03 | 20.80 |
| P. ananatis PA13 | 74.99 | 20.60 |
| P. ananatis VY148 | 74.99 | 20.60 |
| P. ananatis LMG20103 | 74.96 | 20.70 |
| P. eucalypti LMG24197 | 75.89 | 20.90 |
| P. jilinensis D25 | 75.49 | 20.50 |
| P. vagans C9-1 | 75.91 | 20.90 |
| P. vagans FAAARGOS160 | 75.67 | 20.60 |
| P. vagans LMG24199 | 76.02 | 20.90 |
Table 3 OrthoANIu and dDDH scores between strain CCBC3-3-1 and 12 strains of Pantoea
| 物种 Species | CCBC3-3-1 | |
|---|---|---|
| OrthoANIu/% | dDDH/% | |
| P. agglomerans TH81 | 75.95 | 20.90 |
| P. agglomerans C410P1 | 75.66 | 21.00 |
| P. agglomerans CHTF15 | 75.93 | 20.80 |
| P. agglomerans CQ10 | 76.03 | 20.80 |
| P. ananatis PA13 | 74.99 | 20.60 |
| P. ananatis VY148 | 74.99 | 20.60 |
| P. ananatis LMG20103 | 74.96 | 20.70 |
| P. eucalypti LMG24197 | 75.89 | 20.90 |
| P. jilinensis D25 | 75.49 | 20.50 |
| P. vagans C9-1 | 75.91 | 20.90 |
| P. vagans FAAARGOS160 | 75.67 | 20.60 |
| P. vagans LMG24199 | 76.02 | 20.90 |
| 编号 Cluster ID | 类型 Cluster type | 起始-终止位点 Start-stop site | 已知基因簇 Known cluster | 相似度 Similarity/% | 来源 Resources |
|---|---|---|---|---|---|
| 基因簇1 Cluster 1 | 其他(磷酸盐) Other(Phosphonate) | 1 233863 - 1 251863 | - | - | - |
| 基因簇2 Cluster 2 | 糖化物 Saccharide | 1 593 081 - 1 618 729 | O-抗原 O-antigen | 14 | Pseudomonas aeruginosa |
| 基因簇3 Cluster 3 | 核糖体肽类似酶 RiPP-like | 1 674 964 - 1 735 767 | - | - | - |
| 基因簇4 Cluster 4 | 萜烯 Terpene | 2 149 356 - 2 241 512 | 类胡萝卜素 Carotenoid | 100 | Enterobacteriaceae bacterium DC413 |
| 基因簇5 Cluster 5 | 非核糖体多肽合成酶 NRPS | 2 335 269 - 2 383 545 | - | - | - |
| 基因簇6 Cluster 6 | 非核糖体多肽合成酶 NRPS | 3 112 137 - 3 156 045 | Amonabactin | 57 | Aeromonas hydrophila subsp. hydrophila ATCC 7966 |
| 基因簇7 Cluster 7 | 核糖体肽类似酶 RiPP-like | 3 734 150 - 3 762 973 | - | - | - |
| 基因簇8 Cluster 8 | 核糖体肽类似酶 RiPP-like | 4 182 791 - 4 203 075 | - | - | - |
Table 4 Analysis of antibiotic and secondary metabolite gene clusters of strain CCBC3-3-1
| 编号 Cluster ID | 类型 Cluster type | 起始-终止位点 Start-stop site | 已知基因簇 Known cluster | 相似度 Similarity/% | 来源 Resources |
|---|---|---|---|---|---|
| 基因簇1 Cluster 1 | 其他(磷酸盐) Other(Phosphonate) | 1 233863 - 1 251863 | - | - | - |
| 基因簇2 Cluster 2 | 糖化物 Saccharide | 1 593 081 - 1 618 729 | O-抗原 O-antigen | 14 | Pseudomonas aeruginosa |
| 基因簇3 Cluster 3 | 核糖体肽类似酶 RiPP-like | 1 674 964 - 1 735 767 | - | - | - |
| 基因簇4 Cluster 4 | 萜烯 Terpene | 2 149 356 - 2 241 512 | 类胡萝卜素 Carotenoid | 100 | Enterobacteriaceae bacterium DC413 |
| 基因簇5 Cluster 5 | 非核糖体多肽合成酶 NRPS | 2 335 269 - 2 383 545 | - | - | - |
| 基因簇6 Cluster 6 | 非核糖体多肽合成酶 NRPS | 3 112 137 - 3 156 045 | Amonabactin | 57 | Aeromonas hydrophila subsp. hydrophila ATCC 7966 |
| 基因簇7 Cluster 7 | 核糖体肽类似酶 RiPP-like | 3 734 150 - 3 762 973 | - | - | - |
| 基因簇8 Cluster 8 | 核糖体肽类似酶 RiPP-like | 4 182 791 - 4 203 075 | - | - | - |
| 序号No. | 名称Name | MS2分值MS2 score |
|---|---|---|
| 1 | 依诺沙星Enoxacin | 0.99 |
| 2 | 新生霉素Novobiocin | 0.94 |
| 3 | 链脲霉素Streptozocin | 0.85 |
| 4 | 手霉素A Manumycin A | 0.68 |
| 5 | 司帕沙星Sparfloxacin | 0.67 |
| 6 | 阿克拉霉素Aclarubicin | 0.65 |
Table 5 Antibiotics in the fermentation broth of CCBC3-3-1
| 序号No. | 名称Name | MS2分值MS2 score |
|---|---|---|
| 1 | 依诺沙星Enoxacin | 0.99 |
| 2 | 新生霉素Novobiocin | 0.94 |
| 3 | 链脲霉素Streptozocin | 0.85 |
| 4 | 手霉素A Manumycin A | 0.68 |
| 5 | 司帕沙星Sparfloxacin | 0.67 |
| 6 | 阿克拉霉素Aclarubicin | 0.65 |
| [1] | 李海龙, 李端亮. 黄栌属植物研究进展[J]. 陕西林业科技, 2009(6): 22-27. |
| Li HL, Li DL. Advances in studies on genus Cotinus(tourn.) mill[J]. Shaanxi For Sci Technol, 2009(6): 22-27. | |
| [2] | 葛雨萱, 周肖红, 刘洋, 等. 黄栌属种质资源、栽培繁殖、化学成分、叶色调控研究进展[J]. 园艺学报, 2014, 41(9): 1833-1845. |
| Ge YX, Zhou XH, et al. Recent advances in germplasm, cultivation, propagation, chemical components and leaf color regulation of Cotinus[J]. Acta Hortic Sin, 2014, 41(9): 1833-1845. | |
| [3] | 雷增普. 北京地区黄栌黄萎病研究初报[J]. 森林病虫通讯, 1991, 10(3): 12. |
| Lei ZP. Preliminary report on Verticillium wilt of cotinum coggygria in Beijing area[J]. For Pest Dis, 1991, 10(3): 12. | |
| [4] | 雷增普. 北京地区黄栌黄萎病病原菌的研究[J]. 北京林业大学学报, 1993, 15(3): 88-93. |
| Lei ZP. Study of Verticillium causing Cotinus coggygria wilt in the Beijing area[J]. J Beijing For Univ, 1993, 15(3): 88-93. | |
| [5] | Dung JKS, Hamm PB, Eggers JE, et al. Incidence and impact of Verticillium dahliae in soil associated with certified potato seed lots[J]. Phytopathology, 2013, 103(1): 55-63. |
| [6] | 张芸. 棉花内生真菌对棉花黄萎病的控制作用及机理研究[D]. 杨凌: 西北农林科技大学, 2016. |
| Zhang Y. Control effects and mechanism research of cotton endophytic fungi against Verticillium wilt in Gossypium hirsutum[D]. Yangling: Northwest A & F University, 2016. | |
| [7] | 刘璐. 对节白蜡内生细菌YZU-SG146对棉花黄萎病生防作用机制的研究[D]. 荆州: 长江大学, 2021. |
| Liu L. Study on the biological control mechanism of endophytic bacteria YZU-SG146 from Fraxinus hupehensis against Verticillium wilt of cotton[D]. Jingzhou: Yangtze University, 2021. | |
| [8] | 王建美, 田呈明, 过颂新, 等. 黄栌根内生真菌分离鉴定及拮抗真菌筛选[J]. 菌物研究, 2008, 6(1): 35-39. |
| Wang JM, Tian CM, Guo SX, et al. Isolation and screening of endophytic antifungal fungi from Cotinus coggygria roots[J]. J Fungal Res, 2008, 6(1): 35-39. | |
| [9] | 孙妍, 于地美, 等. 黄栌枯萎病菌拮抗细菌的分离与鉴定[J]. 南京林业大学学报: 自然科学版, 2015, 39(6): 17-23. |
| Sun Y, Yu DM, et al. Isolation and identification of an antagonistic bacterium against smoke tree wilt fungus Verticillium dahliae[J]. J Nanjing For Univ Nat Sci Ed, 2015, 39(6): 17-23. | |
| [10] | 许芳. 植物内生细菌对大丽轮枝菌毒素减毒作用的研究[D]. 南京: 南京师范大学, 2012. |
| Xu F. Study on the attenuation of Verticillium dahliae toxin by endophytic bacteria in plants[D]. Nanjing: Nanjing Normal University, 2012. | |
| [11] | Tanizawa Y, Fujisawa T, Kaminuma E, et al. DFAST and DAGA: web-based integrated genome annotation tools and resources[J]. Biosci Microbiota Food Health, 2016, 35(4): 173-184. |
| [12] |
Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication[J]. Bioinformatics, 2018, 34(6): 1037-1039.
doi: 10.1093/bioinformatics/btx713 pmid: 29106469 |
| [13] |
Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition[J]. Proc Natl Acad Sci USA, 2009, 106(45): 19126-19131.
doi: 10.1073/pnas.0906412106 pmid: 19855009 |
| [14] | Wayne LG, Moore WEC, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics[J]. Int J Syst Evol Microbiol, 1987, 37(4): 463-464. |
| [15] | Yoon SH, Ha SM, Lim J, et al. A large-scale evaluation of algorithms to calculate average nucleotide identity[J]. Antonie Van Leeuwenhoek, 2017, 110(10): 1281-1286. |
| [16] | Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, et al. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes[J]. Nucleic Acids Res, 2022, 50(D1): D801-D807. |
| [17] |
Darling ACE, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements[J]. Genome Res, 2004, 14(7): 1394-1403.
doi: 10.1101/gr.2289704 pmid: 15231754 |
| [18] | Blin K, Shaw S, Kloosterman AM, et al. antiSMASH 6.0: improving cluster detection and comparison capabilities[J]. Nucleic Acids Res, 2021, 49(W1): W29-W35. |
| [19] | Reddy BVB, Kallifidas D, Kim JH, et al. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes[J]. Appl Environ Microbiol, 2012, 78(10): 3744-3752. |
| [20] | Gontang EA, Gaudêncio SP, Fenical W, et al. Sequence-based analysis of secondary-metabolite biosynthesis in marine Actinobacteria[J]. Appl Environ Microbiol, 2010, 76(8): 2487-2499. |
| [21] |
Dunn WB, Broadhurst D, Begley P, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry[J]. Nat Protoc, 2011, 6(7): 1060-1083.
doi: 10.1038/nprot.2011.335 pmid: 21720319 |
| [22] |
Kuhl C, Tautenhahn R, Böttcher C, et al. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets[J]. Anal Chem, 2012, 84(1): 283-289.
doi: 10.1021/ac202450g pmid: 22111785 |
| [23] | Zhou JH, Xia F, Che SC, et al. Complete genome sequence of Pantoea sp. strain CCBC3-3-1, an antagonistic endophytic bacterium isolated from a Cotinus coggygria branch[J]. Microbiol Resour Announc, 2019, 8(38):e01004-19. |
| [24] |
Walterson AM, Stavrinides J. Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae[J]. FEMS Microbiol Rev, 2015, 39(6): 968-984.
doi: 10.1093/femsre/fuv027 pmid: 26109597 |
| [25] | Jiang LM, Jeong JC, Lee JS, et al. Potential of Pantoea dispersa as an effective biocontrol agent for black rot in sweet potato[J]. Sci Rep, 2019, 9(1): 16354. |
| [26] |
Smits THM, Duffy B, Blom J, et al. Pantocin A, a peptide-derived antibiotic involved in biological control by plant-associated Pantoea species[J]. Arch Microbiol, 2019, 201(6): 713-722.
doi: 10.1007/s00203-019-01647-7 pmid: 30868174 |
| [27] | Giddens SR, Feng YJ, Mahanty HK. Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola Eh1087[J]. Mol Microbiol, 2002, 45(3): 769-783. |
| [28] |
Pusey PL, Stockwell VO, Reardon CL, et al. Antibiosis activity of Pantoea agglomerans biocontrol strain E325 against Erwinia amylovora on apple flower stigmas[J]. Phytopathology, 2011, 101(10): 1234-1241.
doi: 10.1094/PHYTO-09-10-0253 pmid: 21679036 |
| [29] | Wright SA, Zumoff CH, Schneider L, et al. Pantoea agglomerans strain EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro[J]. Appl Environ Microbiol, 2001, 67(1): 284-292. |
| [30] | Aydin M, Lucht N, König WA, et al. Structure elucidation of the peptide antibiotics herbicolin A and B[J]. Liebigs Ann Chem, 1985(11): 2285-2300. |
| [31] | Vanneste JL, Cornish DA, Yu J, et al. The peptide antibiotic produced by Pantoea agglomerans eh252 is a microcin[J]. Acta Hortic, 2002(590): 285-290. |
| [32] | Vanneste JL, Yu J, Cornish DA. Presence of genes homologous to those necessary for synthesis of microcin mcceh252 in strains of Pantoea agglomerans[J]. Acta Hortic, 2008(793): 391-396. |
| [33] | Xu SD, Liu YX, Cernava T, et al. Fusarium fruiting body microbiome member Pantoea agglomerans inhibits fungal pathogenesis by targeting lipid rafts[J]. Nat Microbiol, 2022, 7(6): 831-843. |
| [1] | TIAN Tong-tong, GE Jia-zhen, GAO Peng-cheng, LI Xue-rui, SONG Guo-dong, ZHENG Fu-ying, CHU Yue-feng. Whole Genome Sequencing and Bioinformatics Analysis of Mycoplasma ovipneumoniae GH3-3 Strain [J]. Biotechnology Bulletin, 2024, 40(7): 323-334. |
| [2] | XU Wei-fang, LI He-yu, ZHANG Hui, HE Zi-ang, GAO Wen-heng, XIE Zi-yang, WANG Chuan-wen, YIN Deng-ke. Efficacy and Its Mechanism of Bacterial Strain HX0037 on the Control of Anthracnose Disease of Trichosanthes kirilowii Maxim [J]. Biotechnology Bulletin, 2024, 40(4): 228-241. |
| [3] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [4] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [5] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [6] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| [7] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [8] | ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101 [J]. Biotechnology Bulletin, 2023, 39(10): 292-303. |
| [9] | ZHANG Ling, ZHANG Rong-yi, LIU Sheng-ke, TAN Zhi-qiong. Screening of Antagonistic Bacteria for Bacterial Fruit Blotch of Cucurbits and Its Antibacterial Effects [J]. Biotechnology Bulletin, 2023, 39(1): 253-263. |
| [10] | WANG Shuai, LV Hong-rui, ZHANG Hao, WU Zhan-wen, XIAO Cui-hong, SUN Dong-mei. Whole-Genome Sequencing Identification of Phosphate-solubilizing Bacteria PSB-R and Analysis of Its Phosphate-solubilizing Properties [J]. Biotechnology Bulletin, 2023, 39(1): 274-283. |
| [11] | WEN Chang, LIU Chen, LU Shi-yun, XU Zhong-bing, AI Chao-fan, LIAO Han-peng, ZHOU Shun-gui. Biological Characteristics and Genome Analysis of a Novel Multidrug-resistant Shigella flexneri Phage [J]. Biotechnology Bulletin, 2022, 38(9): 127-135. |
| [12] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [13] | ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9 [J]. Biotechnology Bulletin, 2022, 38(10): 216-225. |
| [14] | CHEN Ti-qiang, XU Xiao-lan, SHI Lin-chun, ZHONG Li-Yi. Sequencing and Analysis of the Whole Genome of Zizhi Cultivar ‘Wuzhi No.2’(Ganoderma sp. strain Zizhi S2) [J]. Biotechnology Bulletin, 2021, 37(11): 42-56. |
| [15] | GUO He-bao, WANG Xing, HE Shan-wen, ZHANG Xiao-xia. Phenotypic Characteristics Combined with Genomic Analysis to Identify Different Colony Morphology Bacillus velezensis ACCC 19742 [J]. Biotechnology Bulletin, 2020, 36(2): 142-148. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||