








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 63-73.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0205
Previous Articles Next Articles
WU Shuai1( ), XIN Yan-ni1, MAI Chun-hai1, MU Xiao-ya1, WANG Min1, YUE Ai-qin1, ZHAO Jin-zhong3, WU Shen-jie2, DU Wei-jun1(
), XIN Yan-ni1, MAI Chun-hai1, MU Xiao-ya1, WANG Min1, YUE Ai-qin1, ZHAO Jin-zhong3, WU Shen-jie2, DU Wei-jun1( ), WANG Li-xiang1(
), WANG Li-xiang1( )
)
Received:2024-03-04
Online:2024-08-26
Published:2024-09-05
Contact:
DU Wei-jun, WANG Li-xiang
E-mail:1925531945@qq.com;duweijun68@126.com;wanglixiang2@163.com
WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family[J]. Biotechnology Bulletin, 2024, 40(8): 63-73.
| 基因ID Gene ID | 正向引物 Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| GmCYP2 | CGGGACCAGTGTGCTTCTTCA | CCCCTCCACTACAAAGGCTCG |
| GmGS1 | TAACACCAAACAGAGTCATTCACC | TGAGGTTGATGAGATCCGAAA |
| GmGS2 | GACTAATTTCCGGGGTTTCG | GGAGACCTTTTTCTTCCTCACAG |
| GmGS3 | GCTTTTCTTAGTAGGATTTGGTCTC | TAACAATCGGAAAACGAGGGA |
| GmGS4 | GTGGAAGCCATGAGCAAAACT | CGAGGGAAAGGAATAGAAAACA |
| GmGS5 | TTGGAAACCATAAGCAGCCTC | GCCAAGCATTGAAGTGTGAGA |
| GmGS6 | GCAACGTCAAAACAATCACATG | AACAACAGGCGAGGTAGTCACA |
| GmGS7 | GCTGGTGTTGTGCTCTCTCT | TCTTCCCACACGGATTGAGC |
| GmGS8 | ACCCAAAGGTCCAAGCAG | CCAGGATAGCCACCAACG |
Table 1 Primer sequence
| 基因ID Gene ID | 正向引物 Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| GmCYP2 | CGGGACCAGTGTGCTTCTTCA | CCCCTCCACTACAAAGGCTCG |
| GmGS1 | TAACACCAAACAGAGTCATTCACC | TGAGGTTGATGAGATCCGAAA |
| GmGS2 | GACTAATTTCCGGGGTTTCG | GGAGACCTTTTTCTTCCTCACAG |
| GmGS3 | GCTTTTCTTAGTAGGATTTGGTCTC | TAACAATCGGAAAACGAGGGA |
| GmGS4 | GTGGAAGCCATGAGCAAAACT | CGAGGGAAAGGAATAGAAAACA |
| GmGS5 | TTGGAAACCATAAGCAGCCTC | GCCAAGCATTGAAGTGTGAGA |
| GmGS6 | GCAACGTCAAAACAATCACATG | AACAACAGGCGAGGTAGTCACA |
| GmGS7 | GCTGGTGTTGTGCTCTCTCT | TCTTCCCACACGGATTGAGC |
| GmGS8 | ACCCAAAGGTCCAAGCAG | CCAGGATAGCCACCAACG |
| 基因名称 Gene name | 基因座位ID Gene locus ID | 转录本ID Transcript ID | 氨基酸数量Number of amino acids/aa | 理论等电点Theoretical pI | 分子量Molecular weight/kD | 不稳定系Instability index | 亲水平均数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| GmGS1 | Glyma.09g173200 | KRH39032 | 356 | 5.22 | 39212.02 | 40.29 | -0.467 | Cytoplasmic |
| GmGS2 | Glyma.07g104500 | KRH48675 | 356 | 5.32 | 39148 | 40.49 | -0.442 | Cytoplasmic |
| GmGS3 | Glyma.11g215500 | KRH30930 | 356 | 5.48 | 38990.93 | 37.96 | -0.374 | Cytoplasmic |
| GmGS4 | Glyma.18g041100 | KRG97948 | 356 | 5.48 | 39140.98 | 39.47 | -0.425 | Cytoplasmic |
| GmGS5 | Glyma.14g213300 | KRH17324 | 356 | 6.12 | 39208.29 | 38.63 | -0.444 | Cytoplasmic |
| GmGS6 | Glyma.02g244000 | KRH72976 | 356 | 6.03 | 39326.47 | 39.75 | -0.428 | Cytoplasmic |
| GmGS7 | Glyma.13g210800 | KRH48676 | 432 | 6.42 | 47653.72 | 42.55 | -0.466 | Chloroplast |
| GmGS8 | Glyma.15g102000 | KRH30931 | 432 | 6.73 | 47691.83 | 43.16 | -0.458 | Chloroplast |
Table 2 Physical and chemical properties analysis of GS genes in G. max
| 基因名称 Gene name | 基因座位ID Gene locus ID | 转录本ID Transcript ID | 氨基酸数量Number of amino acids/aa | 理论等电点Theoretical pI | 分子量Molecular weight/kD | 不稳定系Instability index | 亲水平均数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| GmGS1 | Glyma.09g173200 | KRH39032 | 356 | 5.22 | 39212.02 | 40.29 | -0.467 | Cytoplasmic |
| GmGS2 | Glyma.07g104500 | KRH48675 | 356 | 5.32 | 39148 | 40.49 | -0.442 | Cytoplasmic |
| GmGS3 | Glyma.11g215500 | KRH30930 | 356 | 5.48 | 38990.93 | 37.96 | -0.374 | Cytoplasmic |
| GmGS4 | Glyma.18g041100 | KRG97948 | 356 | 5.48 | 39140.98 | 39.47 | -0.425 | Cytoplasmic |
| GmGS5 | Glyma.14g213300 | KRH17324 | 356 | 6.12 | 39208.29 | 38.63 | -0.444 | Cytoplasmic |
| GmGS6 | Glyma.02g244000 | KRH72976 | 356 | 6.03 | 39326.47 | 39.75 | -0.428 | Cytoplasmic |
| GmGS7 | Glyma.13g210800 | KRH48676 | 432 | 6.42 | 47653.72 | 42.55 | -0.466 | Chloroplast |
| GmGS8 | Glyma.15g102000 | KRH30931 | 432 | 6.73 | 47691.83 | 43.16 | -0.458 | Chloroplast |
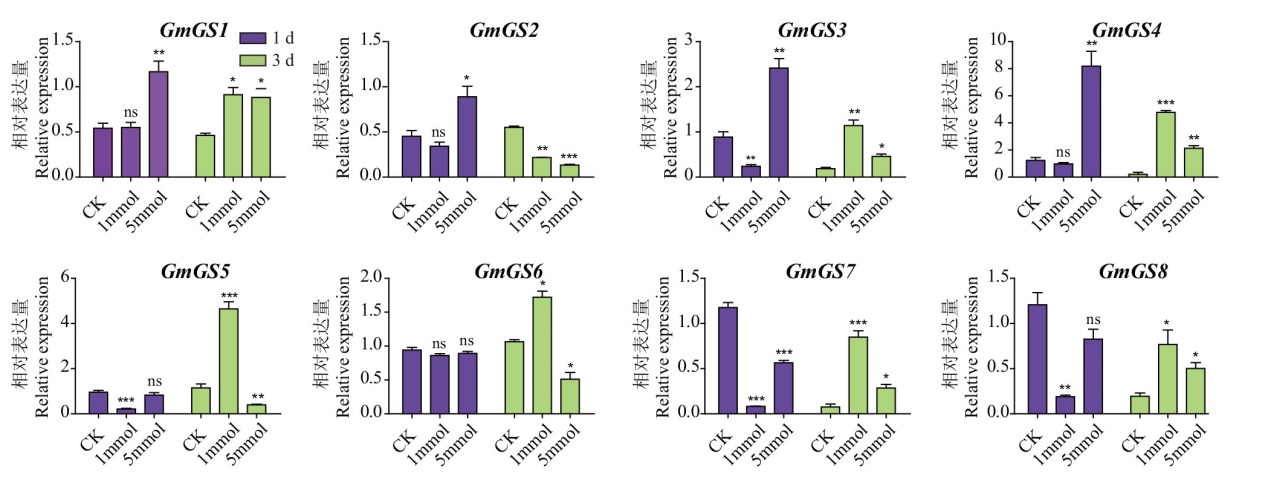
Fig. 7 Expression patterns of G. max GS family genes after ammonium salt treatment 1 d and 3 d indicate 1 and 3 days after treatment. CK, 1 mmol, and 5 mmol are the concentrations of ammonium chloride treatment, representing 0, 1 mmol/L, and 5 mmol/L, respectively. *, **, *** refers to 10%, 5% and 1% significance levels compared to control, and ns to no significant difference. The same below
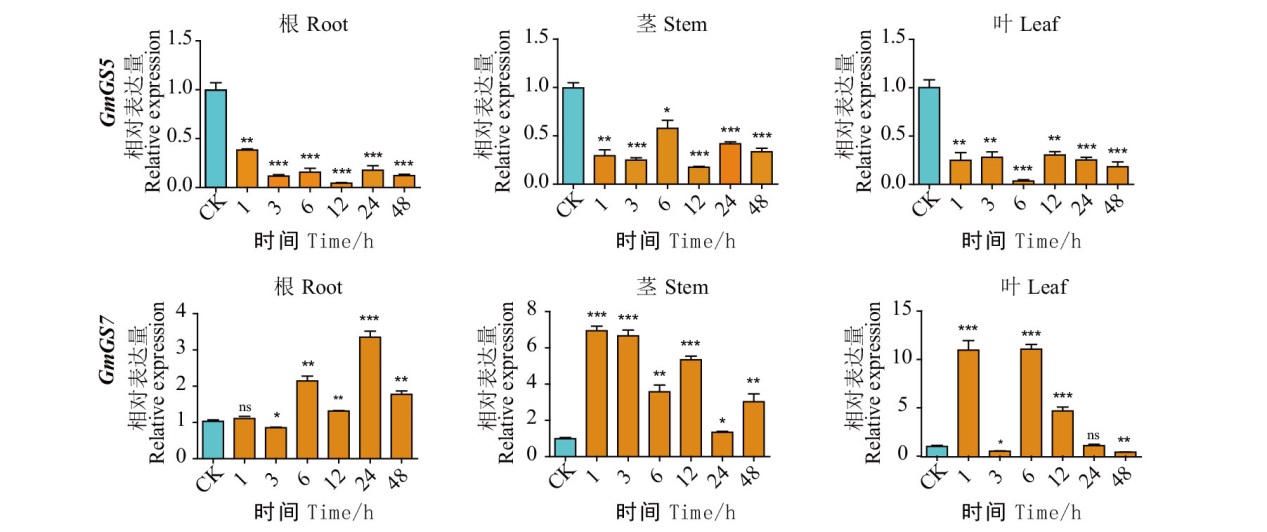
Fig. 8 Expression patterns of G. max GS family genes after salt treatment CK indicates the untreated control, 1, 3, 6, 12, 24, 48 indicate the time after 150 mmol/L NaCI treatment, and the root, stem, and leaf tissue sites of GmGS5 and GmGS7 were sampled from left to right, respectively
| [1] | Zhang Y, Shen SH, Liu YJ. The effect of titanium incorporation on the thermal stability of phenol-formaldehyde resin and its carbonization microstructure[J]. Polym Degrad Stab, 2013, 98(2): 514-518. |
| [2] | 徐洪超, 商靖, 刘铭荟, 等. 氮代谢相关酶的研究进展[J]. 安徽农业科学, 2022, 50(4): 17-20. |
| Xu HC, Shang J, Liu MH, et al. Research progress of enzymes related to nitrogen metabolism[J]. J Anhui Agric Sci, 2022, 50(4): 17-20. | |
| [3] |
Bao AL, Zhao ZQ, Ding GD, et al. The stable level of glutamine synthetase 2 plays an important role in rice growth and in carbon-nitrogen metabolic balance[J]. Int J Mol Sci, 2015, 16(6): 12713-12736.
doi: 10.3390/ijms160612713 pmid: 26053400 |
| [4] | Lee KT, Liao HS, Hsieh MH. Glutamine metabolism, sensing and signaling in plants[J]. Plant Cell Physiol, 2023, 64(12): 1466-1481. |
| [5] | 陈胜勇, 李观康, 汪云, 等. 谷氨酰胺合成酶的研究进展[J]. 中国农学通报, 2010, 26(22): 45-49. |
|
Chen SY, Li GK, Wang Y, et al. The research progress of glutamine synthetase[J]. Chin Agric Sci Bull, 2010, 26(22): 45-49.
doi: 10.11924/j.issn.1000-6850.2010-1942 |
|
| [6] | 韩娜, 葛荣朝, 赵宝存, 等. 植物谷氨酰胺合成酶研究进展[J]. 河北师范大学学报, 2004, 28(4): 407-410, 423. |
| Han N, Ge RC, Zhao BC, et al. Research development of the glutamine synthetase in plants[J]. J Hebei Norm Univ Nat Sci, 2004, 28(4): 407-410, 423. | |
| [7] | 王小纯, 张同勋, 李高飞, 等. 小麦谷氨酰胺合成酶基因克隆与其表达特性分析[J]. 河南农业大学学报, 2012, 46(5): 487-492. |
| Wang XC, Zhang TX, Li GF, et al. Cloning of glutamine synthetases in wheat and analysis of their expression characteristics[J]. J Henan Agric Univ, 2012, 46(5): 487-492. | |
| [8] |
Morey KJ, Ortega JL, Sengupta-Gopalan C. Cytosolic glutamine synthetase in soybean is encoded by a multigene family, and the members are regulated in an organ-specific and developmental manner[J]. Plant Physiol, 2002, 128(1): 182-193.
pmid: 11788764 |
| [9] |
王晓波, 滕婉, 何雪, 等. 大豆谷氨酰胺合成酶基因的分类及根瘤特异表达GmGS1β2基因功能的初步分析[J]. 作物学报, 2013, 39(12): 2145-2153.
doi: 10.3724/SP.J.1006.2013.02145 |
| Wang XB, Teng W, He X, et al. Classification of glutamine synthetase gene and preliminary functional analysis of the nodule-predominantly expressed gene GmGS1β2 in soybean[J]. Acta Agron Sin, 2013, 39(12): 2145-2153. | |
| [10] | 王嘉文, 吴刚, 徐云敏. 谷氨酰胺合成酶在植物氮同化及再利用中的研究进展[J]. 分子植物育种, 2019, 17(4): 1373-1377. |
| Wang JW, Wu G, Xu YM. Research progress of glutamine synthetase in plant nitrogen assimilation and recycling[J]. Mol Plant Breed, 2019, 17(4): 1373-1377. | |
| [11] | 肖急祥, 曾长英, 彭明. 低氮条件下木薯谷氨酰胺合成酶(GS)酶活及GS家族基因表达分析[J]. 分子植物育种, 2016, 14(1): 21-25. |
| Xiao JX, Zeng CY, Peng M. Enzyme activity and expression analysis of glutamine synthetase GS gene in cassava under low-nitrogen stress[J]. Mol Plant Breed, 2016, 14(1): 21-25. | |
| [12] |
Sakakibara H, Shimizu H, Hase T, et al. Molecular identification and characterization of cytosolic isoforms of glutamine synthetase in maize roots[J]. J Biol Chem, 1996, 271(47): 29561-29568.
doi: 10.1074/jbc.271.47.29561 pmid: 8939884 |
| [13] | Caputo C, Criado MV, Roberts IN, et al. Regulation of glutamine synthetase 1 and amino acids transport in the phloem of young wheat plants[J]. Plant Physiol Biochem, 2009, 47(5): 335-342. |
| [14] | 张晓娇. 小麦GS基因家族的鉴定及非生物胁迫响应分析[D]. 郑州: 河南农业大学, 2022. |
| Zhang XJ. Identification and abiotic stress analysis of GS gene family in wheat(Triticum aestivum L.)[D]. Zhengzhou: Henan Agricultural University, 2022. | |
| [15] | 石慧, 王思明. 大豆在中国的历史变迁及其动因探究[J]. 农业考古, 2019(3): 32-39. |
| Shi H, Wang SM. Research on historical development and motivations of soybeans in China[J]. Agric Archaeol, 2019(3): 32-39. | |
| [16] | 张昊, 王文涛. 大豆产业国际竞争力提升的长效机制研究[J]. 湖南农业科学, 2022(6): 81-86. |
| Zhang H, Wang WT. Study on the long-term mechanism of improving the international competitiveness of soybean industry[J]. Hunan Agric Sci, 2022(6): 81-86. | |
| [17] |
石广成, 杨万明, 杜维俊, 等. 大豆耐盐种质的筛选及其耐盐生理特性分析[J]. 生物技术通报, 2022, 38(4): 174-183.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0843 |
| Shi GC, Yang WM, Du WJ, et al. Screening of salt-tolerant soybean germplasm and physiological characteristics analysis of its salt tolerance[J]. Biotechnol Bull, 2022, 38(4): 174-183. | |
| [18] | 王晓丽, 王敏, 岳爱琴, 等. 大豆TGL基因家族全基因组鉴定及高盐胁迫响应研究[J]. 大豆科学, 2022, 41(1): 12-19. |
| Wang XL, Wang M, Yue AQ, et al. Genome-wide identification of soybean TGL gene family and the response to high salt stress[J]. Soybean Sci, 2022, 41(1): 12-19. | |
| [19] |
Gasteiger E, Gattiker A, Hoogland C, et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis[J]. Nucleic Acids Res, 2003, 31(13): 3784-3788.
doi: 10.1093/nar/gkg563 pmid: 12824418 |
| [20] | 陈奕博, 杨万明, 岳爱琴, 等. 大豆LIM转录因子家族鉴定及盐胁迫响应分析[J]. 中国农业大学学报, 2023, 28(4): 26-40. |
| Chen YB, Yang WM, Yue AQ, et al. Identification of soybean LIM transcription factor family and analysis of salt stress response[J]. J China Agric Univ, 2023, 28(4): 26-40. | |
| [21] | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| [22] |
Sun L, Song GS, Guo WJ, et al. Dynamic changes in genome-wide Histone3 Lysine27 trimethylation and gene expression of soybean roots in response to salt stress[J]. Front Plant Sci, 2019, 10: 1031.
doi: 10.3389/fpls.2019.01031 pmid: 31552061 |
| [23] | Bolay P, Muro-Pastor MI, Florencio FJ, et al. The distinctive regulation of cyanobacterial glutamine synthetase[J]. Life, 2018, 8(4): 52. |
| [24] | 姜佳佳. 大豆GmGS1 β2基因及其启动子功能的初步研究[D]. 合肥: 安徽农业大学, 2018. |
| Jiang JJ. Study on function of GmGS1 β2 gene and its promoter in soybean(Glycine max)[D]. Hefei: Anhui Agricultural University, 2018. | |
| [25] |
Castro-Rodríguez V, García-Gutiérrez A, Canales J, et al. The glutamine synthetase gene family in Populus[J]. BMC Plant Biol, 2011, 11: 119.
doi: 10.1186/1471-2229-11-119 pmid: 21867507 |
| [26] | Castro-Rodríguez V, García-Gutiérrez A, Cañas RA, et al. Redundancy and metabolic function of the glutamine synthetase gene family in poplar[J]. BMC Plant Biol, 2015, 15(1): 20. |
| [27] |
Yan Y, Zhang ZH, Sun HW, et al. Nitrate confers rice adaptation to high ammonium by suppressing its uptake but promoting its assimilation[J]. Mol Plant, 2023, 16(12): 1871-1874.
doi: 10.1016/j.molp.2023.11.008 pmid: 37994015 |
| [28] | 李常健, 林清华, 张楚富. 高等植物谷氨酰胺合成酶研究进展[J]. 生物学杂志, 2001, 18(4): 1-3. |
| Li CJ, Lin QH, Zhang CF. Progress of the studies on glutamine synthetase in higher plants[J]. Joural Biol, 2001, 18(4): 1-3. | |
| [29] |
Guan M, Møller IS, Schjoerring JK. Two cytosolic glutamine synthetase isoforms play specific roles for seed germination and seed yield structure in Arabidopsis[J]. J Exp Bot, 2015, 66(1): 203-212.
doi: 10.1093/jxb/eru411 pmid: 25316065 |
| [30] | Konishi N, Ishiyama K, Beier MP, et al. Contributions of two cytosolic glutamine synthetase isozymes to ammonium assimilation in Arabidopsis roots[J]. J Exp Bot, 2017, 68(3): 613-625. |
| [31] | 张春丹, 郑中尧, 刘聪, 等. 拟南芥耐铵突变体的筛选及其AtGLN基因表达分析[J]. 分子植物育种, 2022, 20(18): 6057-6066. |
| Zhang CD, Zheng ZY, Liu C, et al. The screening of high ammonium tolerance mutants of Arabidopsis thaliana and the AtGLN expression analysis[J]. Mol Plant Breed, 2022, 20(18): 6057-6066. | |
| [32] |
Hoshida H, Tanaka Y, Hibino T, et al. Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase[J]. Plant Mol Biol, 2000, 43(1): 103-111.
doi: 10.1023/a:1006408712416 pmid: 10949377 |
| [33] | Debouba M, Gouia H, Suzuki A, et al. NaCl stress effects on enzymes involved in nitrogen assimilation pathway in tomato “Lycopersicon esculentum” seedlings[J]. J Plant Physiol, 2006, 163(12): 1247-1258. |
| [1] | GAO Meng-meng, ZHAO Tian-yu, JIAO Xin-yue, LIN Chun-jing, GUAN Zhe-yun, DING Xiao-yang, SUN Yan-yan, ZHANG Chun-bao. Comparative Transcriptome Analysis of Cytoplasmic Male Sterile Line and Its Restorer Line in Soybean [J]. Biotechnology Bulletin, 2024, 40(7): 137-149. |
| [2] | WANG Fang, YU Lu, QI Ze-zheng, ZHOU Chang-jun, YU Ji-dong. Screening and Biocontrol Effect of Antagonistic Bacteria against Soybean Root Rot [J]. Biotechnology Bulletin, 2024, 40(7): 216-225. |
| [3] | BAI Zhi-yuan, XU Fei, YANG Wu, WANG Ming-gui, YANG Yu-hua, ZHANG Hai-ping, ZHANG Rui-jun. Transcriptome Analysis of Fertility Transformation in Weakly Restoring Hybrid F1 of Soybean Cytoplasmic Male Sterility [J]. Biotechnology Bulletin, 2024, 40(6): 134-142. |
| [4] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [5] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [6] | LI Jing-yan, ZHOU Jia-jing, YUAN Yuan, SU Xiao-yi, QIAO Wen-hui, XUE Yan-lei, LI Guo-jing, WANG Rui-gang. Response of Arabidopsis AtiPGAM2 Gene to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(5): 215-224. |
| [7] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [8] | LI Hui, WEN Yu-fang, WANG Yue, JI Chao, SHI Guo-you, LUO Ying, ZHOU Yong, LI Zhi-min, WU Xiao-yu, YANG You-xin, LIU Jian-ping. Expression Characteristics and Functions of CaPIF4 in Capsicum annuum Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(4): 148-158. |
| [9] | GAO Yu-kun, ZHANG Jian-dong, YANG Pu-yuan, CHEN Dong-ming, WANG Zhi-bo, TIAN Yi-jin, Zakey Eldinn. E. A. Khlid, CUI Jiang-hui, CHANG Jin-hua. Responses of Sorghum Rhizosphere Soil Bacterial Communities to Salt Stress [J]. Biotechnology Bulletin, 2024, 40(4): 203-216. |
| [10] | GAO Zhi-wei, WEI Ming, YU Zu-long, WU Guo-qiang, WEI Jun-long. Identification of Salt-tolerant Plant Growth-promoting Bacterium W-1 and Its Effect on the Salt-tolerance of Sainfoin(Onobrychis viciaefolia) [J]. Biotechnology Bulletin, 2024, 40(4): 217-227. |
| [11] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [12] | SHEN Tian-hong, QI Xiao-bo, ZHAO Rui-feng, MA Xin-rong. Research Progress in the Molecular Mechanisms of Microalgae Responding to Salt Stress [J]. Biotechnology Bulletin, 2024, 40(3): 89-99. |
| [13] | LI Hao, WU Guo-qiang, WEI Ming, HAN Yue-xin. Genome-wide Identification of the BvBADH Gene Family in Sugar Beet(Beta vulgaris)and Their Expression Analysis Under High Salt Stress [J]. Biotechnology Bulletin, 2024, 40(2): 233-244. |
| [14] | XU Yang, ZHANG Rui-ying, DAI Liang-xiang, ZHANG Guan-chu, DING Hong, ZHANG Zhi-meng. Regulation of Nitrogen Application on Peanut Seed Germination and Spermosphere Bacterial Community Structure Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(2): 253-265. |
| [15] | WANG Yu-qing, MA Zi-qi, HOU Jia-xin, ZONG Yu-qi, HAO Han-rui, LIU Guo-yuan, WEI Hui, LIAN Bo-lin, CHEN Yan-hong, ZHANG Jian. Research Progress in the Composition Analysis and Ecological Function of Plant Root Exudates Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(1): 12-23. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||