








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 74-82.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0201
Previous Articles Next Articles
CUI Yuan-yuan( ), WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng(
), WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng( )
)
Received:2024-03-14
Online:2024-08-26
Published:2024-07-30
Contact:
LI Qing-feng
E-mail:18695124679@163.com;liqingfeng2017@nxu.edu.cn
CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage[J]. Biotechnology Bulletin, 2024, 40(8): 74-82.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度Annealing temperature/℃ |
|---|---|---|
| HvNPC1-F | AACAAGCCCCCATCACCG | 58.9 |
| HvNPC1-R | CCACCTTGAAGGCGTCGT | 58.6 |
| HvNPC2-F | ACTACCAGTACCCGCCGT | 58.9 |
| HvNPC2-R | ACCACGTAGTTGGGCAGC | 58.6 |
| HvNPC3-F | GACGTGCTGGACCAGGTC | 59.4 |
| HvNPC3-R | CGGAGGAGAGCAGGGAGT | 59.8 |
| HvNPC4-F | GATGCCGACGAGGATCCG | 59.2 |
| HvNPC4-R | ATCCCCGTTCAGCACTGC | 58.8 |
| HvNPC5-F | CGTCTTCGACCGCTGGTT | 58.4 |
| HvNPC5-R | CGTTGAAGGTGAGCCCGT | 58.6 |
| HvActin-F | CAGGTATCGCTGACCGTATGAG | 57.5 |
| HvActin-R | TGGAAAGTGCTGAGTGAGGCT | 58.9 |
Table 1 Primers used for RT-qPCR
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 退火温度Annealing temperature/℃ |
|---|---|---|
| HvNPC1-F | AACAAGCCCCCATCACCG | 58.9 |
| HvNPC1-R | CCACCTTGAAGGCGTCGT | 58.6 |
| HvNPC2-F | ACTACCAGTACCCGCCGT | 58.9 |
| HvNPC2-R | ACCACGTAGTTGGGCAGC | 58.6 |
| HvNPC3-F | GACGTGCTGGACCAGGTC | 59.4 |
| HvNPC3-R | CGGAGGAGAGCAGGGAGT | 59.8 |
| HvNPC4-F | GATGCCGACGAGGATCCG | 59.2 |
| HvNPC4-R | ATCCCCGTTCAGCACTGC | 58.8 |
| HvNPC5-F | CGTCTTCGACCGCTGGTT | 58.4 |
| HvNPC5-R | CGTTGAAGGTGAGCCCGT | 58.6 |
| HvActin-F | CAGGTATCGCTGACCGTATGAG | 57.5 |
| HvActin-R | TGGAAAGTGCTGAGTGAGGCT | 58.9 |
| 基因名称 Gene name | 基因ID Transcript ID | 物理位置 Physical position | 相对分子量 Molecular weight/kD | 等电点 pI | 亲水性总平均值 Grand average of hydrophilicity | 蛋白长度 Protein length/aa | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| HvNPC1 | HORVU.MOREX.r3.7HG0752530.1 | 7H:631001620-631003350 | 53.12 | 5.22 | -0.317 | 477 | 细胞质 |
| HvNPC2 | HORVU.MOREX.r3.1HG0085590.1 | 1H:496122549-496124474 | 61.33 | 6.53 | -0.364 | 553 | 叶绿体 |
| HvNPC3 | HORVU.MOREX.r3.3HG0232030.1 | 3H:28367740-28369602 | 58.77 | 8.83 | -0.204 | 532 | 叶绿体 |
| HvNPC4 | HORVU.MOREX.r3.3HG0322080.1 | 3H:600541735-600546962 | 59.07 | 5.98 | -0.424 | 533 | 液泡膜 |
| HvNPC5 | HORVU.MOREX.r3.5HG0527960.1 | 5H:566994044-566997667 | 60.76 | 6.80 | -0.367 | 545 | 叶绿体 |
Table 2 Basic physico-chemical properties of NPC family in barley(Hordeum vulgare L)
| 基因名称 Gene name | 基因ID Transcript ID | 物理位置 Physical position | 相对分子量 Molecular weight/kD | 等电点 pI | 亲水性总平均值 Grand average of hydrophilicity | 蛋白长度 Protein length/aa | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|
| HvNPC1 | HORVU.MOREX.r3.7HG0752530.1 | 7H:631001620-631003350 | 53.12 | 5.22 | -0.317 | 477 | 细胞质 |
| HvNPC2 | HORVU.MOREX.r3.1HG0085590.1 | 1H:496122549-496124474 | 61.33 | 6.53 | -0.364 | 553 | 叶绿体 |
| HvNPC3 | HORVU.MOREX.r3.3HG0232030.1 | 3H:28367740-28369602 | 58.77 | 8.83 | -0.204 | 532 | 叶绿体 |
| HvNPC4 | HORVU.MOREX.r3.3HG0322080.1 | 3H:600541735-600546962 | 59.07 | 5.98 | -0.424 | 533 | 液泡膜 |
| HvNPC5 | HORVU.MOREX.r3.5HG0527960.1 | 5H:566994044-566997667 | 60.76 | 6.80 | -0.367 | 545 | 叶绿体 |

Fig. 2 Structural features of NPC gene family in barely A: Purple bars indicate the exon, lines indicate the intron while grey color bars indicate the un-translated region(UTR). B: The yellow bar indicates the signature domain phosphoesterase(PF04185)
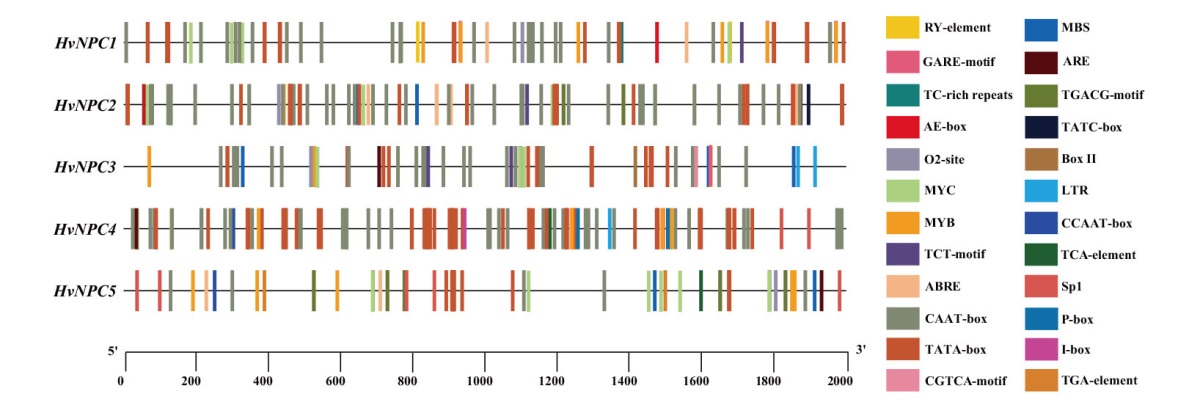
Fig. 3 Cis-acting elements in the promoters of HvNPC gene family members ABRE: Abscisic acid responsive element. CGTCA-motif, TGACG-motif: MeJA responsive element. GARE-motif, P-box, TATC-box: Gibberellin responsive element. TGA-element: Auxin responsive element. TCA-element: Salicylic acid responsive element. MBS, MYC, MYB: Drought responsive element. TC-rich: Defense and stress responsive element. LTR: Low-temperature responsive element. ARE: Essential for the anaerobic responsive element. O2-site: Zein metabolism regulatory element. RY-element: Seed-specific regulatory element. Sp1, TCT-motif, AE-box, BoxII, I-box: Light responsive element
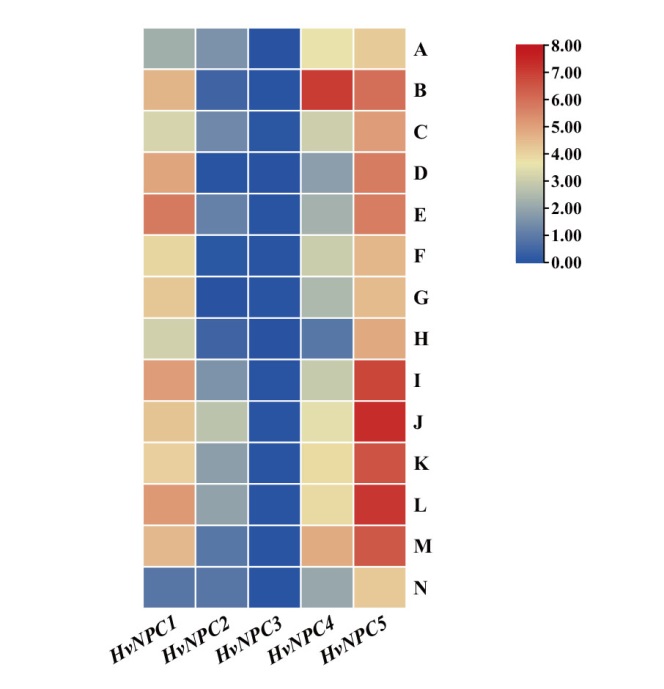
Fig. 4 Expression profile of NPC gene in 14 tissues and organs of barley A: Embryo(4 d after germination). B: Roots from the seedlings(shoot length 10 cm). C: Shoots(10 cm). D: Etiolated seedling (10 d old at dark). E: Developing tillers(six leaf stge, 3rd internode). F: Root(4 weeks). G: Epidermis(4 weeks). H: Florescences(5 mm). I: Florescences(1-1.5 cm). J: Rachis(5 weeks). K: Lemma(6 weeks). L: Lodicule(6 weeks). M: Palea(6 weeks). N: Senescing leaf(2 weeks)
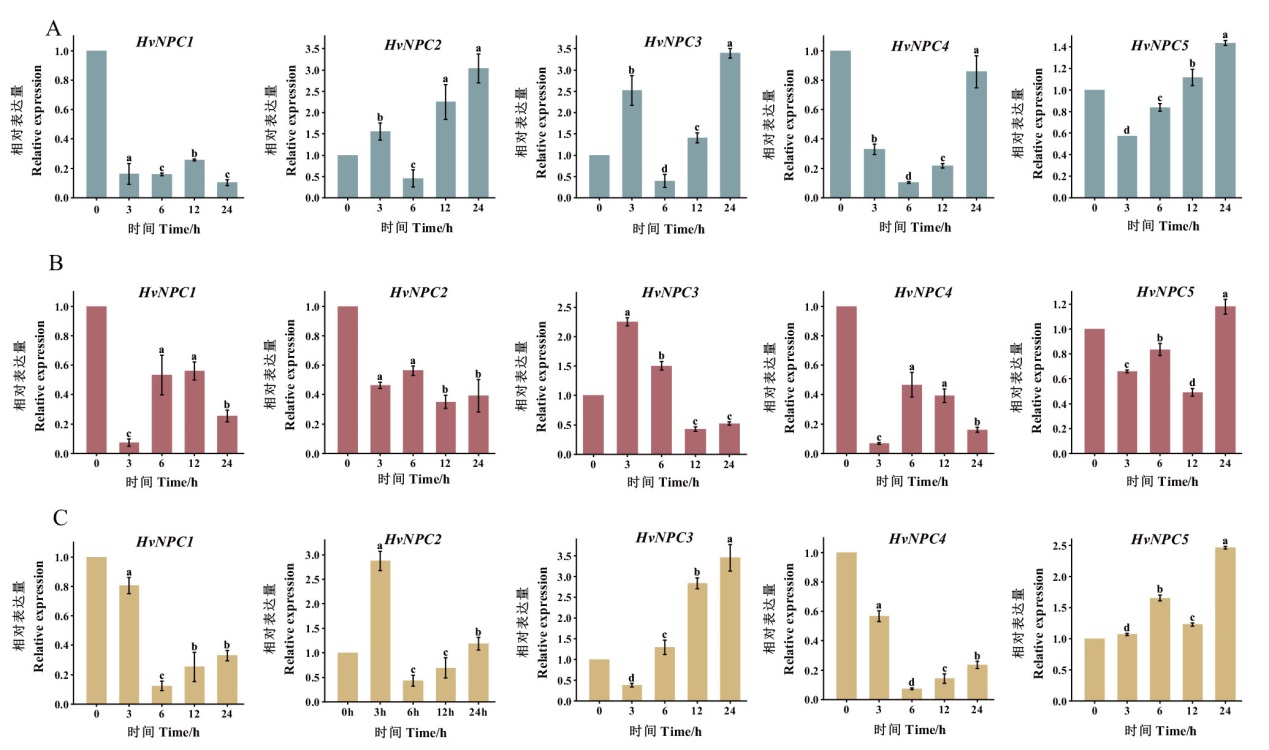
Fig. 5 Relative expression pattern of HvNPCs genes under low temperature, NaCl, and drought treatments A: Gene expression profiles of HvNPCs under 4℃ treatment. B: Gene expression profiles of HvNPCs under NaCl treatment. C: Gene expression profiles of HvNPCs under PEG treatment. Different lower letters indicate significant differences(P<0.05)
| [1] | Zhou J, Wang JJ, Bi YF, et al. Overexpression of PtSOS2 enhances salt tolerance in transgenic poplars[J]. Plant Mol Biol Report, 2014, 32(1): 185-197. |
| [2] |
Wang WX, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance[J]. Planta, 2003, 218(1): 1-14.
doi: 10.1007/s00425-003-1105-5 pmid: 14513379 |
| [3] |
Testerink C, Munnik T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants[J]. J Exp Bot, 2011, 62(7): 2349-2361.
doi: 10.1093/jxb/err079 pmid: 21430291 |
| [4] | Ali U, Lu SP, Fadlalla T, et al. The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses[J]. Prog Lipid Res, 2022, 86: 101158. |
| [5] |
Nakamura Y, Awai K, Masuda T, et al. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis[J]. J Biol Chem, 2005, 280(9): 7469-7476.
doi: 10.1074/jbc.M408799200 pmid: 15618226 |
| [6] | Chen WH, Taylor MC, Barrow RA, et al. Loss of phosphoethanolamine N-methyltransferases abolishes phosphatidylcholine synthesis and is lethal[J]. Plant Physiol, 2019, 179(1): 124-142. |
| [7] | Mou ZL, Wang XQ, Fu ZM, et al. Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis[J]. Plant Cell, 2002, 14(9): 2031-2043. |
| [8] |
Pokotylo I, Pejchar P, Potocký M, et al. The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling[J]. Prog Lipid Res, 2013, 52(1): 62-79.
doi: 10.1016/j.plipres.2012.09.001 pmid: 23089468 |
| [9] |
Krčková Z, Brouzdová J, Daněk M, et al. Arabidopsis non-specific phospholipase C1: characterization and its involvement in response to heat stress[J]. Front Plant Sci, 2015, 6: 928.
doi: 10.3389/fpls.2015.00928 pmid: 26581502 |
| [10] | Krcková Z, Kocourková D, Danek M, et al. The Arabidopsis thaliana non-specific phospholipase C2 is involved in the response to Pseudomonas syringae attack[J]. Ann Bot, 2018, 121(2): 297-310. |
| [11] | Peters C, Li MY, Narasimhan R, et al. Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis[J]. Plant Cell, 2010, 22(8): 2642-2659. |
| [12] | Pejchar P, Potocký M, Krčková Z, et al. Non-specific phospholipase C4 mediates response to aluminum toxicity in Arabidopsis thaliana[J]. Front Plant Sci, 2015, 6: 66. |
| [13] | Peters C, Kim SC, Devaiah S, et al. Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis[J]. Plant Cell Environ, 2014, 37(9): 2002-2013. |
| [14] |
Cai GQ, Fan CC, Liu S, et al. Nonspecific phospholipase C6 increases seed oil production in oilseed Brassicaceae plants[J]. New Phytol, 2020, 226(4): 1055-1073.
doi: 10.1111/nph.16473 pmid: 32176333 |
| [15] | She KJ, Pan WQ, Yan Y, et al. Genome-wide identification, evolution and expressional analysis of OSCA gene family in barley(Hordeum vulgare L.)[J]. Int J Mol Sci, 2022, 23(21): 13027. |
| [16] |
Wang G, Ryu S, Wang X. Plant phospholipases: an overview[J]. Methods Mol Biol, 2012, 861: 123-137.
doi: 10.1007/978-1-61779-600-5_8 pmid: 22426716 |
| [17] | Wang XG, Liu Y, Li Z, et al. Genome-wide identification and expression profile analysis of the phospholipase C gene family in wheat(Triticum aestivum L.)[J]. Plants, 2020, 9(7): 885. |
| [18] | Singh A, Kanwar P, Pandey A, et al. Comprehensive genomic analysis and expression profiling of phospholipase C gene family during abiotic stresses and development in rice[J]. PLoS One, 2013, 8(4): e62494. |
| [19] | Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal[J]. Nucleic Acids Res, 2012, 40(Web Server issue): W597-W603. |
| [20] |
Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Mol Biol Evol, 2011, 28(10): 2731-2739.
doi: 10.1093/molbev/msr121 pmid: 21546353 |
| [21] |
Letunic I, Bork P. Interactive Tree Of Life(iTOL)v4: recent updates and new developments[J]. Nucleic Acids Res, 2019, 47(W1): W256-W259.
doi: 10.1093/nar/gkz239 |
| [22] | Chen CJ, Chen H, He YH, et al. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface[J]. bioRxiv, 2018. DOI: 10.1101/289660. |
| [23] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327.
doi: 10.1093/nar/30.1.325 pmid: 11752327 |
| [24] | Li TT, Pan WQ, Yuan YY, et al. Identification, characterization, and expression profile analysis of the mTERF gene family and its role in the response to abiotic stress in barley(Hordeum vulgare L.)[J]. Front Plant Sci, 2021, 12: 684619. |
| [25] |
Zhang B, Wang YM, Liu JY. Genome-wide identification and characterization of phospholipase C gene family in cotton(Gossypium spp.)[J]. Sci China Life Sci, 2018, 61(1): 88-99.
doi: 10.1007/s11427-017-9053-y pmid: 28547583 |
| [26] | Tasma IM, Brendel V, Whitham SA, et al. Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana[J]. Plant Physiol Biochem, 2008, 46(7): 627-637. |
| [27] | Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis[J]. PLoS Biol, 2004, 2(9): E258. |
| [28] | Zhu JT, Zhou YY, Li JL, et al. Genome-wide investigation of the phospholipase C gene family in Zea mays[J]. Front Genet, 2021, 11: 611414. |
| [29] | Song JL, Zhou YH, Zhang JR, et al. Structural, expression and evolutionary analysis of the non-specific phospholipase C gene family in Gossypium hirsutum[J]. BMC Genomics, 2017, 18(1): 979. |
| [30] | Yang D, Liu X, Yin XM, et al. Rice non-specific phospholipase C6 is involved in mesocotyl elongation[J]. Plant Cell Physiol, 2021, 62(6): 985-1000. |
| [1] | LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui [J]. Biotechnology Bulletin, 2024, 40(8): 174-185. |
| [2] | LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L. [J]. Biotechnology Bulletin, 2024, 40(8): 221-231. |
| [3] | LI Yong-hui, BAO Xing-xing, DUAN Yi-ke, ZHAO Yun-xia, YU Xiang-li, CHEN Yao, ZHANG Yan-zhao. Genome-wide Identification and Expression Features Analysis of the bZIP Family in Rhododendron henanense subsp. lingbaoense [J]. Biotechnology Bulletin, 2024, 40(8): 186-198. |
| [4] | LIU Dan-dan, WANG Lei-gang, SUN Ming-hui, JIAO Xiao-yu, WU Qiong, WANG Wen-jie. Genome-wide Identification and Expression Pattern Profiling of the Trehalose-6-phosphate Synthase(TPS)Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2024, 40(8): 152-163. |
| [5] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [6] | YU Niu, LIU Fan, YANG Jin-chang. Cloning of SgTPS7 in Sindora glabra and Its Function in Terpene Synthesis and Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(8): 164-173. |
| [7] | ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis [J]. Biotechnology Bulletin, 2024, 40(8): 320-328. |
| [8] | ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum [J]. Biotechnology Bulletin, 2024, 40(7): 150-162. |
| [9] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [10] | WU Ding-jie, CHEN Ying-ying, XU Jing, LIU Yuan, ZHANG Hang, LI Rui-li. Research Progress in Plant Gibberellin Oxidase and Its Functions [J]. Biotechnology Bulletin, 2024, 40(7): 43-54. |
| [11] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [12] | ALIYA Waili, CHEN Yong-kun, KELAREMU Kelimujiang, WANG Bao-qing, CHEN Ling-na. Phylogenetic Evolution and Expression Analysis of SPL Gene Family in Juglans regia [J]. Biotechnology Bulletin, 2024, 40(6): 180-189. |
| [13] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [14] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [15] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||