








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (2): 331-342.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0090
LI Wei-hua1( ), WU Jing1, JIN Xue-qin2, LEI Yan-li1(
), WU Jing1, JIN Xue-qin2, LEI Yan-li1( )
)
Received:2024-01-23
Online:2025-02-26
Published:2025-02-28
Contact:
LEI Yan-li
E-mail:546240233@qq.com;912567436@qq.com
LI Wei-hua, WU Jing, JIN Xue-qin, LEI Yan-li. Exploring the Relative Differential Protein Expression of Carbon Tetrachloride-induced Acute Liver Injury in Mice Based on the Proteomics Method[J]. Biotechnology Bulletin, 2025, 41(2): 331-342.

Fig. 1 Liver function-related serum biochemical indexes and liver ultrasound sections in the comparison groupsA, B: The liver parenchyma echo in the comparison group. C: Changes in the maximum oblique position diameter of the right liver in the comparison group. D-H: The expressions of ALT, AST, ALP, GGT, and TBA in the comparison groups. *P<0.05, **P<0.01, ***P<0.001

Fig. 2 CCl4 induction caused gross morphological and histopathological changes (200×)A, B: Plot of gross liver morphology in the comparison group. C: Liver index statistics in the comparison group (n=6). D-E: H & E staining of liver tissue in the comparison groups. F-G: The Masson staining of liver sections's in the comparison groups

Fig. 3 Results of differential protein identification and the GO annotation plots of the differential proteins in comparison groupA: Volcano map of the significant difference in the comparison group. B: GO functional enrichment plots of comparison group differential proteins under BP, CC, and MF (-log10 (P-value)>8.0) biological process classification. C: GO terms plots of the top five up regulated proteins in the comparison group. D: GO terms plots of the top five down regulated proteins in the comparison group
调节(上调/下调) Regulation(Up/Down) | 蛋白编号Accession | 功能描述 Description | 基因符号Gene symbol | 分子量Mw/kD | 比值(CCl4/Control) Ratio(CCl4/Control) | P值 P-value |
|---|---|---|---|---|---|---|
| Up | P50236 | Bile salt sulfotransferase 2 | Sult2a2 | 33.3 | 13.086 | 1.71E-04 |
| Q64449 | C-type mannose receptor 2 | Mrc2 | 167 | 5.077 | 3.11E-04 | |
| P13745 | Glutathione S-transferase A1 | Gsta1 | 25.6 | 4.575 | 2.36E-03 | |
| Q9R100 | Cadherin-17 | Cdh17 | 91.6 | 3.976 | 2.38E-04 | |
| P97501 | Dimethylaniline monooxygenase [N-oxide-forming] 3 | Fmo3 | 60.5 | 3.438 | 1.25E-03 | |
| Q6IS41 | Solute carrier family 25 member 47 | Slc25a47 | 33.6 | 3.224 | 2.49E-02 | |
| Q9CZS1 | Aldehyde dehydrogenase X, mitochondrial | Aldh1b1 | 57.5 | 3.108 | 1.66E-05 | |
| Q8BG95 | Protein phosphatase 1 regulatory subunit 12B | Ppp1r12b | 109 | 3.007 | 1.19E-02 | |
| Down | Q9QXS8 | Probable N-acetyltransferase CML5 | Cml5 | 25.7 | 0.089 | 2.98E-03 |
| Q61694 | NADPH-dependent 3-keto-steroid reductase Hsd3b5 | Hsd3b5 | 41.9 | 0.167 | 1.24E-03 | |
| Q63836 | Selenium-binding protein 2 | Selenbp2 | 52.6 | 0.156 | 4.23E-04 | |
| Q60991 | Cytochrome P450 7B1 | Cyp7b1 | 58.4 | 0.169 | 1.52E-04 | |
| Q9Z0N2 | Eukaryotic translation initiation factor 2 subunit 3, Y-linked | Eif2s3y | 51.1 | 0.21 | 6.87E-05 | |
| O35728 | Cytochrome P450 4A14 | Cyp4a14 | 58.7 | 0.221 | 1.30E-03 | |
| Q9QXZ6 | Solute carrier organic anion transporter family member 1A1 | Slco1a1 | 74.3 | 0.25 | 3.72E-04 | |
| Q61646 | Haptoglobin | Hp | 38.7 | 0.354 | 1.78E-02 | |
| P46425 | Glutathione S-transferase P 2 | Gstp2 | 23.5 | 0.393 | 1.37E-03 | |
| Q3UP75 | UDP-glucuronosyltransferase 3A1 | Ugt3a1 | 59.7 | 0.394 | 5.09E-04 |
Table 1 The top ten up-and down-regulated differential proteins of CCl4-induced liver injury
调节(上调/下调) Regulation(Up/Down) | 蛋白编号Accession | 功能描述 Description | 基因符号Gene symbol | 分子量Mw/kD | 比值(CCl4/Control) Ratio(CCl4/Control) | P值 P-value |
|---|---|---|---|---|---|---|
| Up | P50236 | Bile salt sulfotransferase 2 | Sult2a2 | 33.3 | 13.086 | 1.71E-04 |
| Q64449 | C-type mannose receptor 2 | Mrc2 | 167 | 5.077 | 3.11E-04 | |
| P13745 | Glutathione S-transferase A1 | Gsta1 | 25.6 | 4.575 | 2.36E-03 | |
| Q9R100 | Cadherin-17 | Cdh17 | 91.6 | 3.976 | 2.38E-04 | |
| P97501 | Dimethylaniline monooxygenase [N-oxide-forming] 3 | Fmo3 | 60.5 | 3.438 | 1.25E-03 | |
| Q6IS41 | Solute carrier family 25 member 47 | Slc25a47 | 33.6 | 3.224 | 2.49E-02 | |
| Q9CZS1 | Aldehyde dehydrogenase X, mitochondrial | Aldh1b1 | 57.5 | 3.108 | 1.66E-05 | |
| Q8BG95 | Protein phosphatase 1 regulatory subunit 12B | Ppp1r12b | 109 | 3.007 | 1.19E-02 | |
| Down | Q9QXS8 | Probable N-acetyltransferase CML5 | Cml5 | 25.7 | 0.089 | 2.98E-03 |
| Q61694 | NADPH-dependent 3-keto-steroid reductase Hsd3b5 | Hsd3b5 | 41.9 | 0.167 | 1.24E-03 | |
| Q63836 | Selenium-binding protein 2 | Selenbp2 | 52.6 | 0.156 | 4.23E-04 | |
| Q60991 | Cytochrome P450 7B1 | Cyp7b1 | 58.4 | 0.169 | 1.52E-04 | |
| Q9Z0N2 | Eukaryotic translation initiation factor 2 subunit 3, Y-linked | Eif2s3y | 51.1 | 0.21 | 6.87E-05 | |
| O35728 | Cytochrome P450 4A14 | Cyp4a14 | 58.7 | 0.221 | 1.30E-03 | |
| Q9QXZ6 | Solute carrier organic anion transporter family member 1A1 | Slco1a1 | 74.3 | 0.25 | 3.72E-04 | |
| Q61646 | Haptoglobin | Hp | 38.7 | 0.354 | 1.78E-02 | |
| P46425 | Glutathione S-transferase P 2 | Gstp2 | 23.5 | 0.393 | 1.37E-03 | |
| Q3UP75 | UDP-glucuronosyltransferase 3A1 | Ugt3a1 | 59.7 | 0.394 | 5.09E-04 |
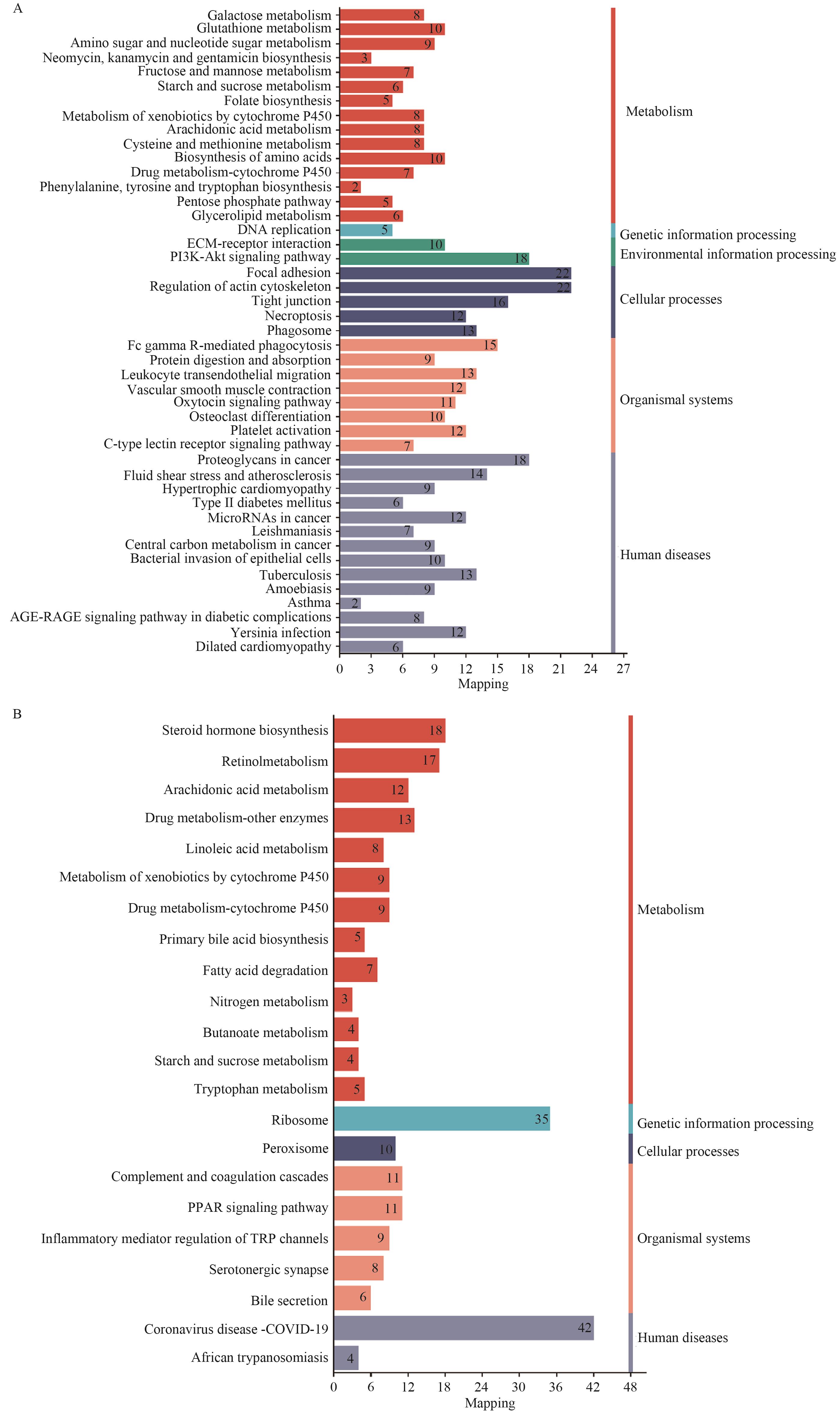
Fig. 4 KEGG pathway classification and functional enrichment plots for up-regulated and down-regulated pathwaysA: Up-regulated pathway. B: Down-regulated pathway
疾病 Diseases | 相关的蛋白 Related proteins | 调节(上调/下调) Regulation(Up/Down) | 费希尔精确检验P值Fisher's exact test P value |
|---|---|---|---|
| Proteoglycans in cancer | P05480 Q63844 P51885 P20444 P49710 Q8BG95 P28654 P70336 P49817 P15379 Q01149 Q8BTM8 P70227 P26041 Q9JKF1 P26040 P11276 P11087 | Up | 7.58E-04 |
| Fluid shear stress and atherosclerosis | Q64337 P05480 P70236 P10648 P16460 Q09014 Q05144 Q64669 P61957 Q5EG47 P49817 Q99L20 P13745 Q60875 | Up | 1.45E-03 |
| Hypertrophic cardiomyopathy | Q6IRU2 P58771 Q60675 P82347 O54950 Q5EG47 A2ARA8 P31001 P09470 | Up | 1.53E-03 |
| MicroRNAs in cancer | P20152 P54227 Q63844 P58771 P63280 P20444 D3Z7P3 P21447 Q64261 P15379 P26645 P26040 | Up | 4.36E-03 |
| Tuberculosis | P05480 Q64449 Q63844 P04441 P19973 Q07813 P51437 O70370 P18242 P11835 O89053 P05555 P08508 | Up | 6.42E-03 |
| Leishmaniasis | Q63844 P28667 Q09014 P11835 Q61093 P05555 P08508 | Up | 5.11E-03 |
| Type II diabetes mellitus | Q63844 P52480 Q91W97 O08528 P28867 P17710 | Up | 2.66E-03 |
| C-type lectin receptor signaling pathway | Q9WVL2 P05480 Q63844 P19973 Q9WTK5 P28867 P70227 | Up | 3.56E-02 |
| Coronavirus disease-COVID-19 | P62918 O09167 P62852 P62849 P62267 P47915 P27659 P61514 P47964 P14115 P62754 P83882 Q9D8E6 P67984 P62830 P62281 Q9CPR4 Q6ZWV7 Q8BP67 P62301 P62751 P41105 P47963 Q9D823 Q9JJI8 P12970 P61255 P61358 Q6ZWV3 P47911 P84099 P62900 Q9D1R9 Q8K182 Q01279 Q8K0E8 Q8VCM7 Q8VCG4 E9PV24 P01029 Q8BH35 P62855 | Down | 1.72E-20 |
| African trypanosomiasis | Q00623 P02088 P02089 P01942 | Down | 1.11E-02 |
Table 2 Related diseases in KEGG analysis
疾病 Diseases | 相关的蛋白 Related proteins | 调节(上调/下调) Regulation(Up/Down) | 费希尔精确检验P值Fisher's exact test P value |
|---|---|---|---|
| Proteoglycans in cancer | P05480 Q63844 P51885 P20444 P49710 Q8BG95 P28654 P70336 P49817 P15379 Q01149 Q8BTM8 P70227 P26041 Q9JKF1 P26040 P11276 P11087 | Up | 7.58E-04 |
| Fluid shear stress and atherosclerosis | Q64337 P05480 P70236 P10648 P16460 Q09014 Q05144 Q64669 P61957 Q5EG47 P49817 Q99L20 P13745 Q60875 | Up | 1.45E-03 |
| Hypertrophic cardiomyopathy | Q6IRU2 P58771 Q60675 P82347 O54950 Q5EG47 A2ARA8 P31001 P09470 | Up | 1.53E-03 |
| MicroRNAs in cancer | P20152 P54227 Q63844 P58771 P63280 P20444 D3Z7P3 P21447 Q64261 P15379 P26645 P26040 | Up | 4.36E-03 |
| Tuberculosis | P05480 Q64449 Q63844 P04441 P19973 Q07813 P51437 O70370 P18242 P11835 O89053 P05555 P08508 | Up | 6.42E-03 |
| Leishmaniasis | Q63844 P28667 Q09014 P11835 Q61093 P05555 P08508 | Up | 5.11E-03 |
| Type II diabetes mellitus | Q63844 P52480 Q91W97 O08528 P28867 P17710 | Up | 2.66E-03 |
| C-type lectin receptor signaling pathway | Q9WVL2 P05480 Q63844 P19973 Q9WTK5 P28867 P70227 | Up | 3.56E-02 |
| Coronavirus disease-COVID-19 | P62918 O09167 P62852 P62849 P62267 P47915 P27659 P61514 P47964 P14115 P62754 P83882 Q9D8E6 P67984 P62830 P62281 Q9CPR4 Q6ZWV7 Q8BP67 P62301 P62751 P41105 P47963 Q9D823 Q9JJI8 P12970 P61255 P61358 Q6ZWV3 P47911 P84099 P62900 Q9D1R9 Q8K182 Q01279 Q8K0E8 Q8VCM7 Q8VCG4 E9PV24 P01029 Q8BH35 P62855 | Down | 1.72E-20 |
| African trypanosomiasis | Q00623 P02088 P02089 P01942 | Down | 1.11E-02 |
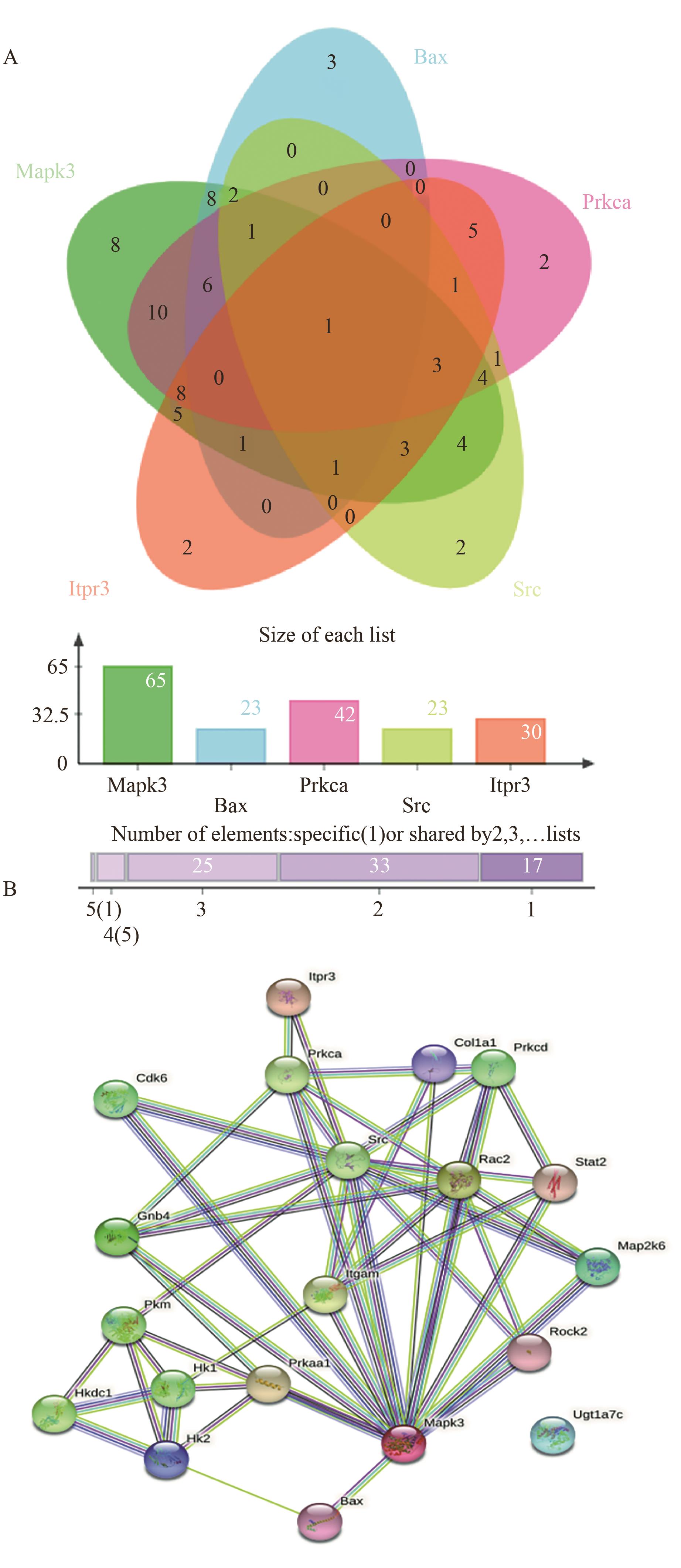
Fig. 5 Protein interaction network diagramA: Degree of differential significant protein ligation Venn plot. B: DAPs network interaction map for comparison group
| 1 | Lachenmeier DW, Monakhova YB, Rehm J. Influence of unrecorded alcohol consumption on liver cirrhosis mortality [J]. World J Gastroenterol, 2014, 20(23): 7217-7222. |
| 2 | Okazaki I, Noro T, Tsutsui N, et al. Fibrogenesis and carcinogenesis in nonalcoholic steatohepatitis (NASH): involvement of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinase (TIMPs) [J]. Cancers, 2014, 6(3): 1220-1255. |
| 3 | Wang S, Lee Y, Kim J, et al. Potential role of Hedgehog pathway in liver response to radiation [J]. PLoS One, 2013, 8(9): e74141. |
| 4 | Xie GH, Karaca G, Swiderska-Syn M, et al. Cross-talk between Notch and Hedgehog regulates hepatic stellate cell fate in mice [J]. Hepatology, 2013, 58(5): 1801-1813. |
| 5 | Yilmaz Y, Eren F. Serum biomarkers of fibrosis and extracellular matrix remodeling in patients with nonalcoholic fatty liver disease: association with liver histology [J]. Eur J Gastroenterol Hepatol, 2019, 31(1): 43-46. |
| 6 | Luckey SW, Petersen DR. Activation of Kupffer cells during the course of carbon tetrachloride-induced liver injury and fibrosis in rats [J]. Exp Mol Pathol, 2001, 71(3): 226-240. |
| 7 | Gan DK, Zhang W, Huang CK, et al. Ursolic acid ameliorates CCl4-induced liver fibrosis through the NOXs/ROS pathway [J]. J Cell Physiol, 2018, 233(10): 6799-6813. |
| 8 | Xu GY, Han X, Yuan GX, et al. Screening for the protective effect target of deproteinized extract of calf blood and its mechanisms in mice with CCl4-induced acute liver injury [J]. PLoS One, 2017, 12(7): e0180899. |
| 9 | McCay PB, Lai EK, Poyer JL, et al. Oxygen- and carbon-centered free radical formation during carbon tetrachloride metabolism. Observation of lipid radicals in vivo and in vitro [J]. J Biol Chem, 1984, 259(4): 2135-2143. |
| 10 | Saijou E, Enomoto Y, Matsuda M, et al. Neutrophils alleviate fibrosis in the CCl4-induced mouse chronic liver injury model [J]. Hepatol Commun, 2018, 2(6): 703-717. |
| 11 | Li H, Zhang T, Wang K, et al. MFGE8 protects against CCl4-induced liver injury by reducing apoptosis and promoting proliferation of hepatocytes [J]. J Cell Physiol, 2019, 234(9): 16463-16474. |
| 12 | Zhao LD, Jin YH, Donahue K, et al. Tissue repair in the mouse liver following acute carbon tetrachloride depends on injury-induced Wnt/β-catenin signaling [J]. Hepatology, 2019, 69(6): 2623-2635. |
| 13 | Liu C, Tao Q, Sun MY, et al. Kupffer cells are associated with apoptosis, inflammation and fibrotic effects in hepatic fibrosis in rats [J]. Lab Invest, 2010, 90(12): 1805-1816. |
| 14 | D'Ambrosio R, Aghemo A, Rumi MG, et al. A morphometric and immunohistochemical study to assess the benefit of a sustained virological response in hepatitis C virus patients with cirrhosis [J]. Hepatology, 2012, 56(2): 532-543. |
| 15 | AlSaid M, Mothana R, Raish M, et al. Evaluation of the effectiveness of Piper cubeba extract in the amelioration of CCl4-induced liver injuries and oxidative damage in the rodent model [J]. Biomed Res Int, 2015, 2015: 359358. |
| 16 | Basu S. Carbon tetrachloride-induced lipid peroxidation: eicosanoid formation and their regulation by antioxidant nutrients [J]. Toxicology, 2003, 189(1-2): 113-127. |
| 17 | Park CM, Cha YS, Youn HJ, et al. Amelioration of oxidative stress by dandelion extract through CYP2E1 suppression against acute liver injury induced by carbon tetrachloride in Sprague-Dawley rats [J]. Phytother Res, 2010, 24(9): 1347-1353. |
| 18 | Son G, Iimuro Y, Seki E, et al. Selective inactivation of NF-kappaB in the liver using NF-kappaB decoy suppresses CCl4-induced liver injury and fibrosis [J]. Am J Physiol Gastrointest Liver Physiol, 2007, 293(3): G631-G639. |
| 19 | Tipoe GL, Leung TM, Liong EC, et al. Epigallocatechin-3-gallate (EGCG) reduces liver inflammation, oxidative stress and fibrosis in carbon tetrachloride (CCl4)-induced liver injury in mice [J]. Toxicology, 2010, 273(1-3): 45-52. |
| 20 | Hsieh CW, Ko WC, Ho WJ, et al. Antioxidant and hepatoprotective effects of Ajuga nipponensis extract by ultrasonic-assisted extraction [J]. Asian Pac J Trop Med, 2016, 9(5): 420-425. |
| 21 | Li SQ, Meng HY, Xi SM, et al. The effect of CCl4-induced acute liver injury on the ADAM8 expression in the mice [C]//2012 International Conference on Biomedical Engineering and Biotechnology. May 28-30, 2012: 589-592. |
| 22 | Vladimir-Knežević S, Cvijanović O, Blažeković B, et al. Hepatoprotective effects of Micromeria croatica ethanolic extract against CCl4-induced liver injury in mice [J]. BMC Complement Altern Med, 2015, 15: 233. |
| 23 | Ma JQ, Ding J, Zhang L, et al. Ursolic acid protects mouse liver against CCl4-induced oxidative stress and inflammation by the MAPK/NF-κB pathway [J]. Environ Toxicol Pharmacol, 2014, 37(3): 975-983. |
| 24 | Fu MZ, Yan YC, Su H, et al. Spleen proteome profiling of dairy goats infected with C. pseudotuberculosis by TMT-based quantitative proteomics approach [J]. J Proteomics, 2021, 248: 104352. |
| 25 | Klaas M, Kangur T, Viil J, et al. The alterations in the extracellular matrix composition guide the repair of damaged liver tissue [J]. Sci Rep, 2016, 6: 27398. |
| 26 | Hao YL, Fang HC, Zhao HL, et al. The role of microRNA-1 targeting of MAPK3 in myocardial ischemia-reperfusion injury in rats undergoing sevoflurane preconditioning via the PI3K/Akt pathway [J]. Am J Physiol Cell Physiol, 2018, 315(3): C380-388. |
| 27 | Runge-Morris M, Kocarek TA, Falany CN. Regulation of the cytosolic sulfotransferases by nuclear receptors [J]. Drug Metab Rev, 2013, 45(1): 15-33. |
| 28 | Mueller JW, Idkowiak J, Gesteira TF, et al. Human DHEA sulfation requires direct interaction between PAPS synthase 2 and DHEA sulfotransferase SULT2A1 [J]. J Biol Chem, 2018, 293(25): 9724-9735. |
| 29 | Bennett BJ, de Aguiar Vallim TQ, Wang ZN, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation [J]. Cell Metab, 2013, 17(1): 49-60. |
| 30 | Kovac S, Angelova PR, Holmström KM, et al. Nrf2 regulates ROS production by mitochondria and NADPH oxidase [J]. Biochim Biophys Acta, 2015, 1850(4): 794-801. |
| 31 | Xie YL, Chu JG, Jian XM, et al. Curcumin attenuates lipopolysaccharide/d-galactosamine-induced acute liver injury by activating Nrf2 nuclear translocation and inhibiting NF-kB activation [J]. Biomed Pharmacother, 2017, 91: 70-77. |
| 32 | Wang QQ, Zhang LL, Yuan XD, et al. The relationship between the bcl-2/bax proteins and the mitochondria-mediated apoptosis pathway in the differentiation of adipose-derived stromal cells into neurons [J]. PLoS One, 2016, 11(10): e0163327. |
| 33 | Yang XS, Li Q, Lin X, et al. Mechanism of fibrotic cardiomyopathy in mice expressing truncated Rho-associated coiled-coil protein kinase 1 [J]. FASEB J, 2012, 26(5): 2105-2116. |
| 34 | Yang L, Xiong ZY, Zhang LJ, et al. Tumor Necrosis Factor Alpha Down-regulated Human GSTA1 and GSTA4 Expression Through The NF-κB Signaling Pathway in Human Hepatoma HepG2 Cells [J]. Progress in Biochemistry and Biophysics, 2016, 43(8): 801-809. |
| 35 | Xie X, Peng J, Chang XT, et al. Activation of RhoA/ROCK regulates NF-κB signaling pathway in experimental diabetic nephropathy [J]. Mol Cell Endocrinol, 2013, 369(1-2): 86-97. |
| [1] | CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics [J]. Biotechnology Bulletin, 2024, 40(3): 135-145. |
| [2] | GU Lei, ZHANG Yu-lu, TANG Shang-rui, YU Hao-yue, LI Chen. Development and Application of Mass Spectrometry-based Single-cell Proteomics Technologies [J]. Biotechnology Bulletin, 2024, 40(11): 125-141. |
| [3] | XU Pei-dong, YI Jian-feng, CHEN Di, CHEN Hao, XIE Bing-yan, ZHAO Wen-jun. Progress in the Application of Omics Technology in Biocontrol Bacillus [J]. Biotechnology Bulletin, 2024, 40(10): 208-220. |
| [4] | ZHOU Lu-qi, CUI Ting-ru, HAO Nan, ZHAO Yu-wei, ZHAO Bin, LIU Ying-chao. Application of Chemical Proteomics in Identifying the Molecular Targets of Natural Products [J]. Biotechnology Bulletin, 2023, 39(9): 12-26. |
| [5] | SANG Tian, WANG Peng-cheng. Research Progress in Plant SUMOylation [J]. Biotechnology Bulletin, 2023, 39(3): 1-12. |
| [6] | ZHAO Ming-ming, TANG Yin, GUO Lei-zhou, HAN Jia-hui, GE Jia-ming, MENG Yong, PING Shu-zhen, ZHOU Zheng-fu, WANG Jin. Function Analysis of Lon1 Protease Involved in High Temperature Stress and Cell Division of Deinococcus radiodurans R1 [J]. Biotechnology Bulletin, 2022, 38(5): 149-158. |
| [7] | DING Ya-qun, DING Ning, XIE Shen-min, HUANG Meng-na, ZHANG Yu, ZHANG Qin, JIANG Li. Construction of Vps28 Knock-out Mice and Model Study of the Impact on Lactation and Immune Traits [J]. Biotechnology Bulletin, 2022, 38(3): 164-172. |
| [8] | LI Bing-juan, ZHENG Lu, SHEN Ren-fang, LAN Ping. Proteomic Analysis of RPP1A Involved in the Seedling Growth of Arabidopsis thaliana [J]. Biotechnology Bulletin, 2022, 38(2): 10-20. |
| [9] | LI Zhi-hao, ZHANG Ge, MO Zhi-jie, DENG Shuai-jun, LI Jia-yi, ZHANG Hai-bo, LIU Xiao-hui, LIU Hao-bao. Effects of a Xylanase-producing Bacillus cereus on the Composition and Fermented Products of Cigar Leaves [J]. Biotechnology Bulletin, 2022, 38(2): 105-112. |
| [10] | WANG Zhi-bo, WANG Dao-ping, MIAO Lan, LI Ying, PAN Ying-hong, LIU Jian-xun. Comparative Study on Methods of Analyzing Proteome in Blood Samples [J]. Biotechnology Bulletin, 2021, 37(8): 307-318. |
| [11] | XIE Shao-yi, JIANG Lu-man, YANG Xiao-feng, ZHANG Xian-yu, WU Zhen-fang, LI Zi-cong. Construction and Verification of CRISPR/Cas9 System Expression Vector for Mouse X Chromosome Cutting [J]. Biotechnology Bulletin, 2021, 37(5): 67-75. |
| [12] | MENG Li-ná, PENG Chun-ying, LI Tie-dong, LI Bo-sheng. Proteomic ánálysis of Spiruliná plátensis in Response to ársenic Stress [J]. Biotechnology Bulletin, 2020, 36(4): 107-116. |
| [13] | LI Kun, LIU Yue, HUANG Peng, YANG Zhi-fang, HU Qian, ZHANG Ying, LI Zhi-hong, LÜ Ye-hui, LIANG Le. Proteomics Study on Spermatogonia Differentiation in Mice [J]. Biotechnology Bulletin, 2020, 36(3): 168-176. |
| [14] | WANG Qi-wen, LI Pan, Pan Cui-yun, HAN Fen-xia. Effect of Ethylene Glycol on the Expression of Exogenous Genes in Vivo [J]. Biotechnology Bulletin, 2019, 35(4): 64-68. |
| [15] | ZHANG Liang, CHEN Xiao-qing, SONG Jia-yu, MAO Ran-ran, JIANG Qian-wen, LIN Xiang-min. Comparative Proteomics Analysis of Escherichia coli in Response to Barofloxacin Stress [J]. Biotechnology Bulletin, 2019, 35(3): 103-109. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||