








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (3): 62-70.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0784
Previous Articles Next Articles
LIANG Li-cun1( ), WANG Ke-fen2, SONG Zu-huan1, LIU Meng-ting1, LI Jia-yu1, LUO Hui-ying1, YAO Bin1, YANG Hao-meng1(
), WANG Ke-fen2, SONG Zu-huan1, LIU Meng-ting1, LI Jia-yu1, LUO Hui-ying1, YAO Bin1, YANG Hao-meng1( )
)
Received:2024-08-15
Online:2025-03-26
Published:2025-03-20
Contact:
YANG Hao-meng
E-mail:lianglicun1022@163.com;yhmbjbj@126.com
LIANG Li-cun, WANG Ke-fen, SONG Zu-huan, LIU Meng-ting, LI Jia-yu, LUO Hui-ying, YAO Bin, YANG Hao-meng. Improving the Efficiency of Gene Editing by Optimizing sgRNA in Aspergillus tubingensis[J]. Biotechnology Bulletin, 2025, 41(3): 62-70.
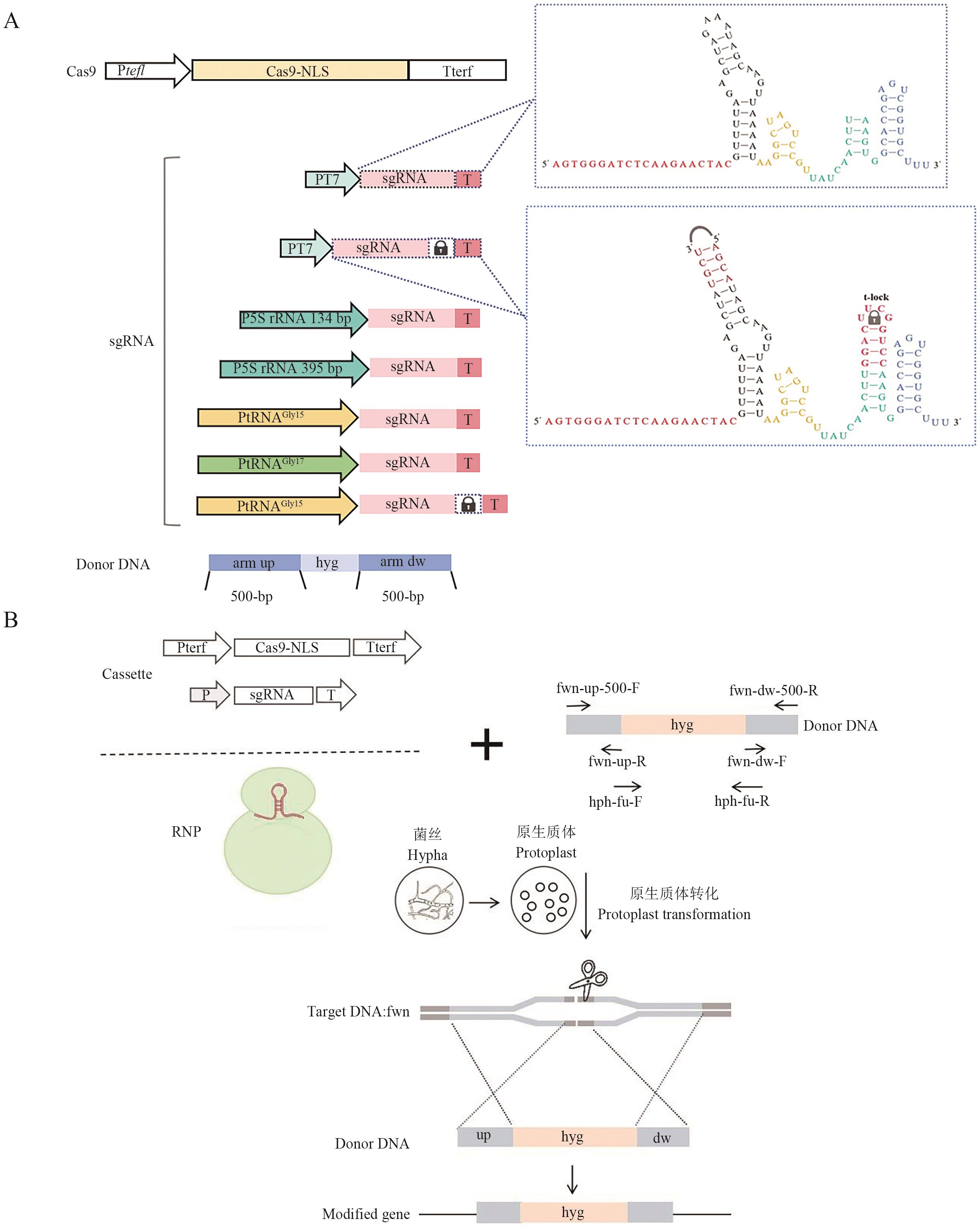
Fig. 1 Schematic diagram of gene editing element structure and editing processA: The left side from top to bottom indicates the composition of cas9 expression cassette, sgRNA expression cassette of different promoters and donor DNA, respectively. The Cas9 expression cassette is composed of Pterf promoter, coding sequence, nuclear localization signal peptide NLS and Tterf terminator. The promoters of sgRNA are T7 promoter, 5SrRNA promoter (including 134 bp and 395 bp), tRNAGly15 promoter and tRNAGly17 promoter, respectively. The tracrRNA skeleton of normal sgRNA is in the blue dotted box above the right side, and the sgRNA tracrRNA skeleton in the dotted box below carries a hairpin structure represented by a “lock” graph. Donor DNA consists of 500 bp upstream and downstream homologous arms and intermediate hygromycin resistance genes. B: A brief schematic diagram of the CRISPR/Cas9 system, which shows the editing originals required for the expression cassette method and the RNP method. When the expression cassette method is used for transformation, the Cas9 expression cassette and sgRNA expression cassette need to be added to the prepared protoplasts together with Donor DNA. When the RNP method was used for transformation, Cas9 protein and sgRNA were incubated in vitro, and the two were added to the prepared protoplasts together with Donor DNA in the form of RNP complex for transformation. The target gene fwnA was cut, and the genome sequences on both sides of the cutting site were homologously recombined with the homologous arm of Donor DNA, thus the hygromycin gene was inserted into the fwnA gene, and the gene was inactivated, resulting in the spore color from black to white, and the gene detection primers were labeled on the schematic diagram of Donor DNA
| 引物名称Primer name | 序列Sequence |
|---|---|
| fwn-up-500-F | 5'-CAACCAAGCTCAAGGTTCCTTACGCG-3' |
| fwn-up-R | 5'-GTTCTTGAGATCCCACTTGTAGGCTGGG-3' |
| fwn-dw-F | 5'-CTATACCAACAACTTCTGCCTGAGCAAGG-3' |
| fwn-dw-500-R | 5'-TCCGCAGCAGCGCAGTCAAAG-3' |
| hph-fu-F | 5'-CAGGCTACAAGTGGGATCTCAAGAACGACAGAAGATGATATTGAAGGAGCACTTTTTG-3' |
| hph-fu-R | 5'-AGTTGTTGGTATAGGGAATCCAGTAGTATCTGGAAGAGGTAAACCCGAAACG-3' |
| 5SRNA-F1 | 5'-ACGAAGAGGATGGTTGAACACGGAT-3' |
| 5SRNA-F2 | 5'-CACATACGACCACAGGGTGTG-3' |
| P5SRNA-R | 5'-ACATACAACAGAAGGGATTCGCTGGTG-3' |
| 5SRNA-overlap-F | 5'-CACCAGCGAATCCCTTCTGTTGTATGTAGTGGGATCTCAAGAACTACGTTTTAGAGC-3' |
| sgRNA-R | 5'-AAAAGCACCGACTCGGTGCC-3' |
| CAS9-F | 5'-CGAGACAGCAGAATCACCGCCCAAGTTAAG-3' |
| CAS9-R | 5'-ATTACACTTGTATTGGGATGAATTTTGTATGCAC-3' |
| tRNAGly15-F | 5'-CTCCGTAGATAGAGATAGGAGTGGATAGGGA-3' |
| tRNAGly15-R | 5'-TGCATCATCCGTGAATCGAACACGG-3' |
| tRNAGly17-F | 5'-ACAATTCACAATCCGCAAGACGTTAACG-3' |
| tRNAGly17-R | 5'-TGCATCATCCGTGAATCGAACACGG-3' |
| tRNAGly15/17-overlap-F | 5'-CCGTGTTCGATTCACGGATGATGCAAGTGGGATCTCAAGAACTACGTTTTAG-3' |
Table 1 Primers used in this study
| 引物名称Primer name | 序列Sequence |
|---|---|
| fwn-up-500-F | 5'-CAACCAAGCTCAAGGTTCCTTACGCG-3' |
| fwn-up-R | 5'-GTTCTTGAGATCCCACTTGTAGGCTGGG-3' |
| fwn-dw-F | 5'-CTATACCAACAACTTCTGCCTGAGCAAGG-3' |
| fwn-dw-500-R | 5'-TCCGCAGCAGCGCAGTCAAAG-3' |
| hph-fu-F | 5'-CAGGCTACAAGTGGGATCTCAAGAACGACAGAAGATGATATTGAAGGAGCACTTTTTG-3' |
| hph-fu-R | 5'-AGTTGTTGGTATAGGGAATCCAGTAGTATCTGGAAGAGGTAAACCCGAAACG-3' |
| 5SRNA-F1 | 5'-ACGAAGAGGATGGTTGAACACGGAT-3' |
| 5SRNA-F2 | 5'-CACATACGACCACAGGGTGTG-3' |
| P5SRNA-R | 5'-ACATACAACAGAAGGGATTCGCTGGTG-3' |
| 5SRNA-overlap-F | 5'-CACCAGCGAATCCCTTCTGTTGTATGTAGTGGGATCTCAAGAACTACGTTTTAGAGC-3' |
| sgRNA-R | 5'-AAAAGCACCGACTCGGTGCC-3' |
| CAS9-F | 5'-CGAGACAGCAGAATCACCGCCCAAGTTAAG-3' |
| CAS9-R | 5'-ATTACACTTGTATTGGGATGAATTTTGTATGCAC-3' |
| tRNAGly15-F | 5'-CTCCGTAGATAGAGATAGGAGTGGATAGGGA-3' |
| tRNAGly15-R | 5'-TGCATCATCCGTGAATCGAACACGG-3' |
| tRNAGly17-F | 5'-ACAATTCACAATCCGCAAGACGTTAACG-3' |
| tRNAGly17-R | 5'-TGCATCATCCGTGAATCGAACACGG-3' |
| tRNAGly15/17-overlap-F | 5'-CCGTGTTCGATTCACGGATGATGCAAGTGGGATCTCAAGAACTACGTTTTAG-3' |
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| PT7-sgRNA-fwn | TAATACGACTCACTATAGGAGTGGGATCTCAAGAACTACGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT |
| PT7-lock-sgRNA-fwn | TAATACGACTCACTATAGGAGTGGGATCTCAAGAACTACGTTTTAGAGCTA |
Table 2 Sequence information of sgRNA in vitro transcription template
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| PT7-sgRNA-fwn | TAATACGACTCACTATAGGAGTGGGATCTCAAGAACTACGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT |
| PT7-lock-sgRNA-fwn | TAATACGACTCACTATAGGAGTGGGATCTCAAGAACTACGTTTTAGAGCTA |

Fig. 2 Amplification of Cas9 expression cassette, sgRNA expression cassette and Donor DNA1: sgRNA with 134 bp 5S rRNA as promoter; 2: sgRNA with 395 bp 5S rRNA as promoter; 3: a sgRNA with tRNAGly17 as the promoter; 4: a sgRNA with tRNAGly15 as the promoter; 5: sgRNA with tRNAGly15 as promoter and fused with “lock” structure; 6: Donor DNA; 7:Cas9 expression cassette

Fig. 3 Growth of transformants and the gene editing efficiency of different sgRNA promoters during transformation by expression cassette methodA: The growths of transformants in the two experimental groups of 5S rRNA 134 bp and tRNAGly15 (the colors of black spores, white spores, and miscellaneous spores are shown in the figure). B: The gene editing efficiency of sgRNA with different lengths of 5S promoter, tRNAGly15 promoter and tRNAGly17 promoter

Fig. 4 Gene editing efficiency of sgRNA with “lock” structure in RNP methodThe histogram in Fig.4 shows the gene editing efficiency of sgRNA containing ordinary sgRNA structure and sgRNA fused with “lock” structure.Three biological replicates were performed in each experimental group, and the obtained data were analyzed by spss software. The error line indicates the size of the standard error.The significance level between different letters is 0.05 ( the same below )
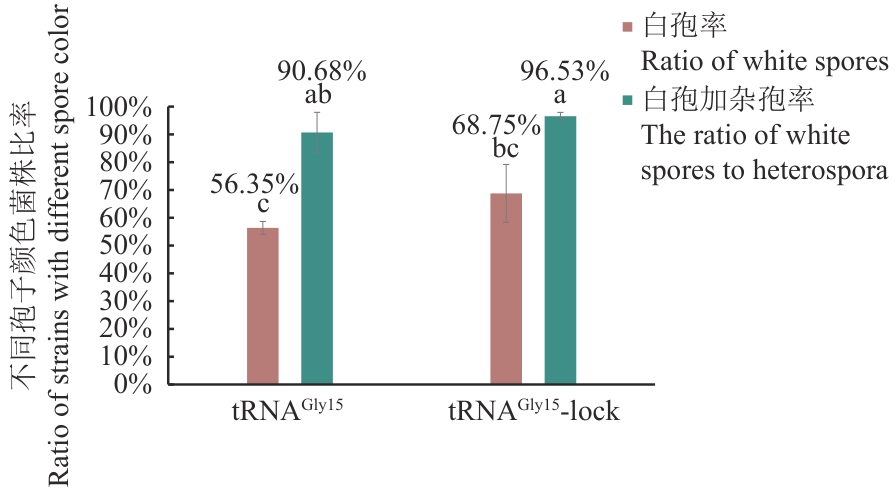
Fig. 5 Gene editing efficiency of tRNAGly15 promoter superimposed with “lock” structure in expression cassette methodThe gene editing efficiency of sgRNA with tRNAGly15 as the promoter and sgRNA with tRNAGly15 as the promoter and fused with “lock” structure
| 1 | Gaur S, Kaur M, Kalra R, et al. Application of microbial resources in biorefineries: current trend and future prospects [J]. Heliyon, 2024, 10(8): e28615. |
| 2 | 塔宾曲霉菌——治理白色污染的又一“利器" [J]. 塑料科技, 2017, 45(7): 57. |
| Aspergillus tabini-another "sharp weapon" to control white pollution [J]. Plast Sci Technol, 2017, 45(7): 57. | |
| 3 | Krisnawati R, Cahyanto MN, Sardjono S, et al. RNA-seq data of Aspergillus tubingensis NBRC 31125 in carbon catabolite repressor related to xylanase production [J]. Data Brief, 2022, 45: 108700. |
| 4 | Huang DM, Song YY, Liu YL, et al. A new strain of Aspergillus tubingensis for high-activity pectinase production [J]. Braz J Microbiol, 2019, 50(1): 53-65. |
| 5 | Okado N, Sugi M, Kasamoto S, et al. Safety evaluation of Arabinase (arabinan endo-1, 5-α-L-arabinanase) from Aspergillus tubingensis [J]. Food Sci Nutr, 2019, 8(1): 456-478. |
| 6 | Olarte RA, Horn BW, Singh R, et al. Sexual recombination in Aspergillus tubingensis [J]. Mycologia, 2015, 107(2): 307-312. |
| 7 | Deng HX, Gao RJ, Liao XR, et al. CRISPR system in filamentous fungi: current achievements and future directions [J]. Gene, 2017, 627: 212-221. |
| 8 | Jin FJ, Wang BT, Wang ZD, et al. CRISPR/Cas9-based genome editing and its application in Aspergillus species [J]. J Fungi, 2022, 8(5): 467. |
| 9 | 肖晗, 刘宜欣. CRISPR-Cas系统编辑丝状真菌的进展与挑战 [J]. 合成生物学, 2021, 2(2): 274-286. |
| Xiao H, Liu YX. Progress and challenge of the CRISPR-Cas system in gene editing for filamentous fungi [J]. Synth Biol J, 2021, 2(2): 274-286. | |
| 10 | Zou G, Xiao ML, Chai SX, et al. Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents [J]. Microb Biotechnol, 2021, 14(6): 2343-2355. |
| 11 | Kubicek CP, Harman GE. Trichoderma and gliocladium volume 1:Basic biology, taxonomy and genetics [M]. London: Taylor and Francis Ltd, 1998. |
| 12 | 杨浩萌, 梁丽存, 宋祖洹, 等. 提高丝状真菌精准基因编辑效率的策略 [J]. 微生物学通报, 2024, 51(5): 1425-1440. |
| Yang HM, Liang LC, Song ZH, et al. Strategies for improving the efficiency of precise gene editing in filamentous fungi [J]. Microbiol China, 2024, 51(5): 1425-1440. | |
| 13 | 高伟欣, 黄火清, 赵晶, 等. 应用于基因编辑的核糖核蛋白复合体的构建与活性验证 [J]. 生物技术通报, 2022, 38(8): 60-68. |
| Gao WX, Huang HQ, Zhao J, et al. Construction and activity verification of ribonucleoprotein complex for gene editing [J]. Biotechnol Bull, 2022, 38(8): 60-68. | |
| 14 | Kuivanen J, Korja V, Holmström S, et al. Development of microtiter plate scale CRISPR/Cas9 transformation method for Aspergillus niger based on in vitro assembled ribonucleoprotein complexes [J]. Fungal Biol Biotechnol, 2019, 6: 3. |
| 15 | Schuster M, Kahmann R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes [J]. Fungal Genet Biol, 2019, 130: 43-53. |
| 16 | Song LT, Ouedraogo JP, Kolbusz M, et al. Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger [J]. PLoS One, 2018, 13(8): e0202868. |
| 17 | Schuster M, Schweizer G, Reissmann S, et al. Genome editing in Ustilago maydis using the CRISPR-Cas system [J]. Fungal Genet Biol, 2016, 89: 3-9. |
| 18 | Liu J, Zhu J, Zhang Q, et al. Establishing a one-step marker-free CRISPR/Cas9 system for industrial Aspergillus niger using counter-selectable marker Ang-ace2 [J]. Biotechnol Lett, 2023, 45(11-12): 1477-1485. |
| 19 | Zheng XM, Zheng P, Sun JB, et al. Heterologous and endogenous U6 snRNA promoters enable CRISPR/Cas9 mediated genome editing in Aspergillus niger [J]. Fungal Biol Biotechnol, 2018, 5: 2. |
| 20 | Zheng XM, Zheng P, Zhang K, et al. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger [J]. ACS Synth Biol, 2019, 8(7): 1568-1574. |
| 21 | Katayama T, Tanaka Y, Okabe T, et al. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae [J]. Biotechnol Lett, 2016, 38(4): 637-642. |
| 22 | Riesenberg S, Helmbrecht N, Kanis P, et al. Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage [J]. Nat Commun, 2022, 13(1): 489. |
| 23 | Dong C, Gou YW, Lian JZ. SgRNA engineering for improved genome editing and expanded functional assays [J]. Curr Opin Biotechnol, 2022, 75: 102697. |
| 24 | Nødvig CS, Hoof JB, Kogle ME, et al. Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli [J]. Fungal Genet Biol, 2018, 115: 78-89. |
| 25 | Zhang T, Gao YB, Wang RC, et al. Production of guide RNAs in vitro and in vivo for crispr using ribozymes and RNA polymerase II promoters [J]. Bio Protoc, 2017, 7(4): e2148. |
| 26 | Wang Q, Zhao QQ, Liu Q, et al. CRISPR/Cas9-mediated genome editing in Penicillium oxalicum and Trichoderma reesei using 5S rRNA promoter-driven guide RNAs [J]. Biotechnol Lett, 2021, 43(2): 495-502. |
| 27 | Fan C, Zhang W, Su XY, et al. CRISPR/Cas9-mediated genome editing directed by a 5S rRNA-tRNAGly hybrid promoter in the thermophilic filamentous fungus Humicola insolens [J]. Biotechnol Biofuels, 2021, 14(1): 206. |
| 28 | Zhang JX, Li KH, Sun Y, et al. An efficient CRISPR/Cas9 genome editing system based on a multiple sgRNA processing platform in Trichoderma reesei for strain improvement and enzyme production [J]. Biotechnol Biofuels Bioprod, 2024, 17(1): 22. |
| 29 | Arazoe T, Miyoshi K, Yamato T, et al. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus [J]. Biotechnol Bioeng, 2015, 112(12): 2543-2549. |
| 30 | Jung WJ, Park SJ, Cha S, et al. Factors affecting the cleavage efficiency of the CRISPR-Cas9 system [J]. Anim Cells Syst, 2024, 28(1): 75-83. |
| 31 | Sarkari P, Marx H, Blumhoff ML, et al. An efficient tool for metabolic pathway construction and gene integration for Aspergillus niger [J]. Bioresour Technol, 2017, 245(Pt B): 1327-1333. |
| 32 | Kuivanen J, Arvas M, Richard P. Clustered genes encoding 2-keto-l-gulonate reductase and l-Idonate 5-Dehydrogenase in the novel fungal d-glucuronic acid pathway [J]. Front Microbiol, 2017, 8: 225. |
| 33 | Shen CC, Hsu MN, Chang CW, et al. Synthetic switch to minimize CRISPR off-target effects by self-restricting Cas9 transcription and translation [J]. Nucleic Acids Res, 2019, 47(3): e13. |
| 34 | Kwon MJ, Schütze T, Spohner S, et al. Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi [J]. Fungal Biol Biotechnol, 2019, 6: 15. |
| [1] | WU Shu-ning, SU Yong-ping, LI Dong-xue, BAI Ying-guo, LIU Bo, ZHANG Zhi-wei. Design and Application of a Cumate-inducible Promoter for Corynebacterium glutamicum [J]. Biotechnology Bulletin, 2024, 40(7): 108-116. |
| [2] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [3] | WANG Zhou, YU Jie, WANG Jin-hua, WANG Yong-ze, ZHAO Xiao. Anaerobic Expression of Lactate Dehydrogenase to Improve the D-lactic Acid Optical Purity Procluced by Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(5): 290-299. |
| [4] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| [5] | LIU Yu-ping, ZHANG Wei-yang, ZHANG Wei-min, YE Wei, LI Dong-li. Identification of the Promoter for the Biosynthesis Gene of Polyketide Meroterpenoids in Phomopsis tersa FS441 [J]. Biotechnology Bulletin, 2024, 40(12): 248-255. |
| [6] | WANG Zi-rui, LIU Xiao-han, XIU Yu, LIN Shan-zhi. Cloning of the Promoter of MATE40 Gene from Prunus sibirica Seeds and Analysis of Gene Expression Driven by Promoter [J]. Biotechnology Bulletin, 2024, 40(11): 192-201. |
| [7] | ZHAO Rui, DI Jing-yi, ZHANG Guang-tong, LIU Hao, GAO Wei-xia. Screening Endogenous Expression Elements in Streptococcus zooepidemicus via Transcriptomics Analysis and Applications for High Yield of Hyaluronic Acid [J]. Biotechnology Bulletin, 2024, 40(10): 296-304. |
| [8] | ZHANG Jie-ping, GUAN Yue-feng. Crop Breeding Based on Promoter Editing [J]. Biotechnology Bulletin, 2024, 40(10): 53-61. |
| [9] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [10] | LIU Yu-ling, WANG Meng-yao, SUN Qi, MA Li-hua, ZHU Xin-xia. Effect of RD29A Promoter on the Stress Resistance of Transgenic Tobacco with SikCDPK1 Gene from Saussurea involucrata [J]. Biotechnology Bulletin, 2023, 39(9): 168-175. |
| [11] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [12] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [13] | SHI Guang-zhen, WANG Zhao-ye, SUN Qi, ZHU Xin-xia. Cloning and Activity Analysis of SikCDPK1 Promoter from Saussurea involucrata [J]. Biotechnology Bulletin, 2022, 38(9): 191-197. |
| [14] | CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. pOsHAK1:OsFLN2 Expression Enhances the Drought Tolerance by Altering Sugar Metabolism in Rice [J]. Biotechnology Bulletin, 2022, 38(8): 92-100. |
| [15] | NIE Li-bin, YI Ling-xin, DENG Yan, SHENG Qi, WU Xiao-yu, ZHANG Bin. Pathway Engineering Modification of Corynebacterium glutamicum for Shikimic Acid Production [J]. Biotechnology Bulletin, 2022, 38(6): 93-102. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||