








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (8): 1-10.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0300
CAI Ru-feng( ), YANG Yu-xuan, YU Ji-zheng, LI Jia-nan(
), YANG Yu-xuan, YU Ji-zheng, LI Jia-nan( )
)
Received:2025-03-20
Online:2025-08-26
Published:2025-08-14
Contact:
LI Jia-nan
E-mail:2363763510@qq.com;lydian_l@163.com
CAI Ru-feng, YANG Yu-xuan, YU Ji-zheng, LI Jia-nan. Artificial Intelligence Transforms Protein Engineering: From Structural Analysis to Synthetic Biology through Algorithmic Advancements[J]. Biotechnology Bulletin, 2025, 41(8): 1-10.
模型 Model | 发布时间 Release time | CASP参赛版本 CASP entry version | GDT_TS中位数 GDT_TS med-number | 覆盖UniProt比例 Coverage ratio of UniProt | 关键技术革新 Key technological innovation |
|---|---|---|---|---|---|
| AlphaFold1 | 2018 | CASP13 | 68.5 | 35% | 残基距离图预测 |
| AlphaFold2 | 2020 | CASP14 | 92.4 | 98% | Evoformer架构 |
| AlphaFold3 | 2024 | - | - | 全结构域 | 多聚体建模 |
Table 1 Comparison of performance across AlphaFold model series
模型 Model | 发布时间 Release time | CASP参赛版本 CASP entry version | GDT_TS中位数 GDT_TS med-number | 覆盖UniProt比例 Coverage ratio of UniProt | 关键技术革新 Key technological innovation |
|---|---|---|---|---|---|
| AlphaFold1 | 2018 | CASP13 | 68.5 | 35% | 残基距离图预测 |
| AlphaFold2 | 2020 | CASP14 | 92.4 | 98% | Evoformer架构 |
| AlphaFold3 | 2024 | - | - | 全结构域 | 多聚体建模 |
| 特征 Characteristic | AlphaFold2 | RoseTTAFold |
|---|---|---|
| 计算资源 | 128 TPU v3 (4 d/蛋白) | 4 GPU (8 h/蛋白) |
| 核心架构 | Evoformer+结构模块 | 三轨Transformer |
| 动态结构预测 | 单构象输出 | 支持构象系综生成 |
| 膜蛋白预测精度 | TM-score 0.72 | TM-score 0.81 |
| 开源程度 | 部分开源 | 全代码公开 |
Table 2 Technical comparison between RoseTTAFold and AlphaFold2
| 特征 Characteristic | AlphaFold2 | RoseTTAFold |
|---|---|---|
| 计算资源 | 128 TPU v3 (4 d/蛋白) | 4 GPU (8 h/蛋白) |
| 核心架构 | Evoformer+结构模块 | 三轨Transformer |
| 动态结构预测 | 单构象输出 | 支持构象系综生成 |
| 膜蛋白预测精度 | TM-score 0.72 | TM-score 0.81 |
| 开源程度 | 部分开源 | 全代码公开 |

Fig. 1 Three-dimensional structures of β2-AR obtained by different methodsA: X-ray method, with PDB ID 4G8R; B: NMR method, with PDB ID 6KR8; C: cryo-EM method, with PDB ID 8GGI; D: AlphaFold prediction result, with the number AF-P07550
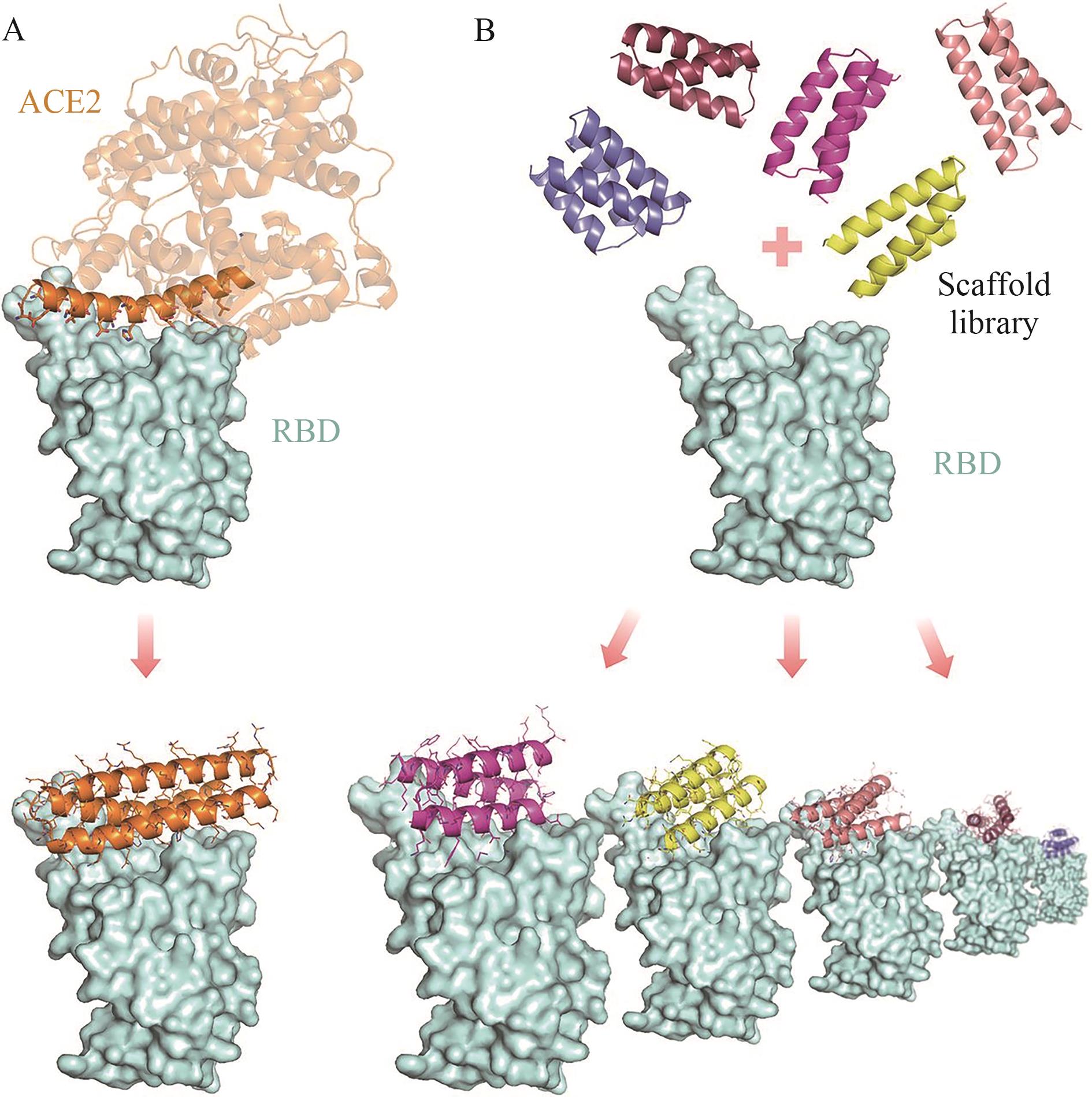
Fig. 2 Overview diagram of computational design methodA: Design of helical proteins containing ACE2 helix. B:Large-scale head-to-tail design of small helical scaffolds followed by RIF docking to identify shape and chemical complementary binding patterns
| [1] | Woolfson DN. A brief history of de novo protein design: minimal, rational, and computational [J]. J Mol Biol, 2021, 433(20): 167160. |
| [2] | Simon I, Magyar C. Assortment of frontiers in protein science [J]. Int J Mol Sci, 2022, 23(7): 3685. |
| [3] | Zhuravlev PI, Papoian GA. Protein functional landscapes, dynamics, allostery: a tortuous path towards a universal theoretical framework [J]. Q Rev Biophys, 2010, 43(3): 295-332. |
| [4] | AlQuraishi M. Machine learning in protein structure prediction [J]. Curr Opin Chem Biol, 2021, 65: 1-8. |
| [5] | Zhou BX, Tan Y, Hu YT, et al. Protein engineering in the deep learning era [J]. mLife, 2024, 3(4): 477-491. |
| [6] | 潘杰, 中山皓博. 人工智能及其在生命科学中的应用与展望 [J]. 山东师范大学学报: 自然科学版, 2024, 39(2): 117-142. |
| Pan J, Akihiro Nakayama. Artificial intelligence and its application and prospect in life sciences [J]. J Shandong Norm Univ Nat Sci Ed, 2024, 39(2): 117-142. | |
| [7] | Gupta V, Liao WK, Choudhary A, et al. Evolution of artificial intelligence for application in contemporary materials science [J]. MRS Commun, 2023, 13(5): 754-763. |
| [8] | Pederson T. Protein structure: has levinthal’s paradox “folded”? [J]. FASEB J, 2021, 35(3): e21416. |
| [9] | Wang NN, Dong J, Ouyang DF. AI-directed formulation strategy design initiates rational drug development [J]. J Control Release, 2025, 378: 619-636. |
| [10] | Senior AW, Evans R, Jumper J, et al. Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13) [J]. Proteins, 2019, 87(12): 1141-1148. |
| [11] | 阎隆飞, 孙之荣. 蛋白质分子结构 [M]. 北京: 清华大学出版社, 1999: 211-213. |
| Yan LF, Sun ZR. Protein molecular structure [M]. Beijing: Tsinghua University Press, 1999: 211-213. | |
| [12] | Berman HM, Battistuz T, Bhat TN, et al. The protein data bank [J]. Nucleic Acids Research, 2000, 28(1): 235-242. |
| [13] | 宁正元, 林世强. 蛋白质结构的预测及其应用 [J]. 福建农业大学学报, 2006, 35(3): 308-313. |
| Ning ZY, Lin SQ. Protein structure prediction and its application [J]. J Fujian Agric For Univ Nat Sci Ed, 2006, 35(3): 308-313. | |
| [14] | 杜宗阳. 蛋白质与RNA三级结构预测算法研究 [D]. 天津: 南开大学, 2022. |
| Du ZY. Research on algorithms for protein and RNA tertiary structure prediction [D]. Tianjin: Nankai University, 2022. | |
| [15] | 张晓凯, 张丛丛, 刘忠民, 等. 冷冻电镜技术的应用与发展 [J]. 科学技术与工程, 2019, 19(24): 9-17. |
| Zhang XK, Zhang CC, Liu ZM, et al. Application and development of cryo-electron microscopy technology [J]. Sci Technol Eng, 2019, 19(24): 9-17. | |
| [16] | 郭贝一, 郭晓强. AlphaFold和蛋白质结构预测 [J]. 科学, 2024, 76(5): 39-44. |
| Guo BY, Guo XQ. AlphaFold and protein structure prediction [J]. Science, 2024, 76(5): 39-44. | |
| [17] | Shaw DE, Maragakis P, Lindorff-Larsen K, et al. Atomic-level characterization of the structural dynamics of proteins [J]. Science, 2010, 330(6002): 341-346. |
| [18] | Lindorff-Larsen K, Piana S, Dror RO, et al. How fast-folding proteins fold [J]. Science, 2011, 334(6055): 517-520. |
| [19] | Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules [J]. Nat Struct Biol, 2002, 9(9): 646-652. |
| [20] | Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold [J]. Nature, 2021, 596(7873): 583-589. |
| [21] | Varadi M, Anyango S, Deshpande M, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models [J]. Nucleic Acids Res, 2022, 50(d1): D439-D444. |
| [22] | Li Y, Zhang CX, Feng CJ, et al. Integrating end-to-end learning with deep geometrical potentials for ab initio RNA structure prediction [J]. Nat Commun, 2023, 14(1): 5745. |
| [23] | Dauparas J, Anishchenko I, Bennett N, et al. Robust deep learning-based protein sequence design using ProteinMPNN [J]. Science, 2022, 378(6615): 49-56. |
| [24] | Baek M, DiMaio F, Anishchenko I, et al. Accurate prediction of protein structures and interactions using a three-track neural network [J]. Science, 2021, 373(6557): 871-876. |
| [25] | Lisanza SL, Gershon JM, Tipps SWK, et al. Multistate and functional protein design using RoseTTAFold sequence space diffusion [J]. Nat Biotechnol, 2024. |
| [26] | Mirdita M, Schütze K, Moriwaki Y, et al. ColabFold: making protein folding accessible to all [J]. Nat Methods, 2022, 19(6): 679-682. |
| [27] | Velloso JPL, Kovacs AS, Pires DEV, et al. AI-driven GPCR analysis, engineering, and targeting [J]. Curr Opin Pharmacol, 2024, 74: 102427. |
| [28] | Singh I, Seth A, Billesbølle CB, et al. Structure-based discovery of conformationally selective inhibitors of the serotonin transporter [J]. Cell, 2023, 186(10): 2160-2175.e17. |
| [29] | Pándy-Szekeres G, Caroli J, Mamyrbekov A, et al. GPCRdb in 2023: state-specific structure models using AlphaFold2 and new ligand resources [J]. Nucleic Acids Res, 2023, 51(d1): D395-D402. |
| [30] | 王子佳, 郭卫娜, 郭巧珍, 等. β2-肾上腺素受体激动剂在神经退行性病变相关认知障碍中的作用 [J]. 中国医学科学院学报, 2022, 44(6): 1112-1116. |
| Wang ZJ, Guo WN, Guo QZ, et al. Role of β2-adrenergic receptor agonist in the cognitive impairment associated with neurodegenerative diseases [J]. Acta Academiae Medicinae Sinicae, 2022, 44(6): 1112-1116. | |
| [31] | Ramos BP, Colgan LA, Nou E, et al. β2 adrenergic agonist, clenbuterol, enhances working memory performance in aging animals [J]. Neurobiol Aging, 2008, 29(7): 1060-1069. |
| [32] | Humphreys IR, Pei J, Baek M, et al. Computed structures of core eukaryotic protein complexes [J]. Science, 2021, 374(6573): eabm4805. |
| [33] | Goverde CA, Wolf B, Khakzad H, et al. De novo protein design by inversion of the AlphaFold structure prediction network [J]. Protein Sci, 2023, 32(6): e4653. |
| [34] | Love AC, Prescher JA. Seeing (and using) the light: recent developments in bioluminescence technology [J]. Cell Chem Biol, 2020, 27(8): 904-920. |
| [35] | Jiang TY, Du LP, Li MY. Lighting up bioluminescence with coelenterazine: strategies and applications [J]. Photochem Photobiol Sci, 2016, 15(4): 466-480. |
| [36] | Yeh AH, Norn C, Kipnis Y, et al. De novo design of luciferases using deep learning [J]. Nature, 2023, 614(7949): 774-780. |
| [37] | Rohl CA, Strauss CEM, Misura KMS, et al. Protein structure prediction using Rosetta [J]. Meth Enzymol, 2004, 383: 66-93. |
| [38] | Dou JY, Vorobieva AA, Sheffler W, et al. De novo design of a fluorescence-activating β-barrel [J]. Nature, 2018, 561(7724): 485-491. |
| [39] | Cao L, Goreshnik I, Coventry B, et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors [J]. Science, 2020, 370(6515): 426-431. |
| [40] | 谭生龙. 基于序列的蛋白质功能分类系统的研究与设计 [J]. 科技创新与应用, 2016, 6(27): 68. |
| Tan SL. Research and design of protein function classification system based on sequence [J]. Technol Innov Appl, 2016, 6(27): 68. | |
| [41] | 叶玉珍, 丁达夫. 蛋白质骨架库的构建及其在功能蛋白质设计中的应用 [J]. 生物物理学报, 1999, 15(4): 751-757. |
| Ye YZ, Ding DF. Construction of protein scaffold database and itsapplications to functional protein design [J]. Acta Biophys Sin, 1999, 15(4): 751-757. | |
| [42] | Li YL, Jiao WT, Liu RH, et al. Expanding the sequence spaces of synthetic binding protein using deep learning-based framework ProteinMPNN [J]. Front Comput Sci, 2024, 19(5): 195903. |
| [43] | Sumida KH, Núñez-Franco R, Kalvet I, et al. Improving protein expression, stability, and function with ProteinMPNN [J]. J Am Chem Soc, 2024, 146(3): 2054-2061. |
| [44] | Watson JL, Juergens D, Bennett NR, et al. De novo design of protein structure and function with RFdiffusion [J]. Nature, 2023, 620(7976): 1089-1100. |
| [45] | Robins K. BPS2025-Enhancing antibody design using RFdiffusion and ProteinMPNN for novel intrabody generation [J]. Biophysical Journal, 2025, 124(3):217a-218a. |
| [46] | Whitesides GM. The origins and the future of microfluidics [J]. Nature, 2006, 442(7101): 368-373. |
| [47] | Snoek J, Larochelle H, Adams PR. Practical bayesian optimization of machine learning algorithms [J]. CoRR, 2012. |
| [48] | Abramson J, Adler J, Dunger J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold3 [J]. Nature, 2024, 630(8016): 493-500. |
| [49] | Nguyen E, Poli M, Durrant MG, et al. Sequence modeling and design from molecular to genome scale with Evo [J]. Science, 2024, 386(6723): eado9336. |
| [50] | 张裕, 周化岚, 张建国, 等. 无细胞蛋白质表达系统的优化与应用 [J]. 生命的化学, 2022, 42(8): 1493-1501. |
| Zhang Y, Zhou HL, Zhang JG, et al. Optimization and application of cell-free system for protein expression [J]. Chem Life, 2022, 42(8): 1493-1501. | |
| [51] | Mazzotti G, Hartmann D, Booth MJ. Precise, orthogonal remote-control of cell-free systems using photocaged nucleic acids [J]. J Am Chem Soc, 2023, 145(17): 9481-9487. |
| [52] | Graham F. Daily briefing: AlphaFold developers share Nobel prize in chemistry [J]. Nature, 2024. |
| [53] | Evseev P, Shneider M, Miroshnikov K. Evolution of phage tail sheath protein [J]. Viruses, 2022, 14(6): 1148. |
| [54] | Podgorski JM, Freeman K, Gosselin S, et al. A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability [J]. Structure, 2023, 31(3): 282-294.e5. |
| [55] | Bisio H, Legendre M, Giry C, et al. Evolution of giant pandoravirus revealed by CRISPR/Cas9 [J]. Nat Commun, 2023, 14: 428. |
| [56] | Bian JH, Tan P, Nie T, et al. Optimizing enzyme thermostability by combining multiple mutations using protein language model [J]. mLife, 2024, 3(4): 492-504. |
| [57] | Liao YJ, Ma H, Wang ZY, et al. Rapid restoration of potent neutralization activity against the latest Omicron variant JN.1 via AI rational design and antibody engineering [J]. Proc Natl Acad Sci USA, 2025, 122(6): e2406659122. |
| [58] | Wu T, Chen XH, Fei YT, et al. Artificial metalloenzyme assembly in cellular compartments for enhanced catalysis [J]. Nat Chem Biol, 2025, 21(5): 779-789. |
| [59] | Jiang F, Li MC, Dong JJ, et al. A general temperature-guided language model to design proteins of enhanced stability and activity [J]. Sci Adv, 2024, 10(48): eadr2641. |
| [1] | HUANG Xu-sheng, ZHOU Ya-li, CHAI Xu-dong, WEN Jing, WANG Ji-ping, JIA Xiao-yun, LI Run-zhi. Cloning of Plastidial PfLPAT1B Gene from Perilla frutescens and Its Functional Analysis in Oil Biosynthesis [J]. Biotechnology Bulletin, 2025, 41(7): 226-236. |
| [2] | GAO Jing, CHENG Yi-cun, GAO Ming, ZHAO Yun-xiao, WANG Yang-dong. Regulation of Plant Tannin Synthesis and Mechanisms of Its Responses to Environment [J]. Biotechnology Bulletin, 2025, 41(7): 49-59. |
| [3] | WANG Hui, FAN Ling-xi, SUN Ji-lu, WANG Yuan, WU Ning-feng, TIAN Jian, GUAN Fei-fei. Enhancing the Thermostability of Lysozyme RPL187 Based on Protein Intelligence Models [J]. Biotechnology Bulletin, 2025, 41(7): 336-346. |
| [4] | WU Ya, YAO Run, YANG Han-ting, LIU Wei, YANG Shuai, SONG Chi, CHEN Shi-lin. Genome-wide Identification and Expression Analysis of SDR Gene Family in Mentha suaveolens ‘Variegata’ [J]. Biotechnology Bulletin, 2025, 41(5): 175-185. |
| [5] | LU Tian-yi, LI Ai-peng, FEI Qiang. Research Progress in the Biosynthesis of Polylactic Acid [J]. Biotechnology Bulletin, 2025, 41(4): 47-60. |
| [6] | LI Xiao-ming, SHANG Xiu-hua, WANG You-shuang, WU Zhi-hua. Research Progress in Benzoxazinoids in Plants [J]. Biotechnology Bulletin, 2025, 41(4): 9-20. |
| [7] | NIE Zhu-xin, GUO Jin, QIAO Zi-yang, LI Wei-wei, ZHANG Xue-yan, LIU Chun-yang, WANG Jing. Transcriptome Analysis of the Anthocyanin Biosynthesis in the Fruit Development Processes of Lycium ruthenicum Murr. [J]. Biotechnology Bulletin, 2024, 40(8): 106-117. |
| [8] | MA Xiao-xiang, MA Ze-yuan, LIU Ya-yue, ZHOU Long-jian, HE Yi-fan, ZHANG Yi. Effects of Simulated Mutational Biosynthetic Regulation on the Secondary Metabolites of Aspergillus terreus C23-3 [J]. Biotechnology Bulletin, 2024, 40(8): 275-287. |
| [9] | SHEN Zhen-hui, CAO Yao, YANG Lin-lei, LUO Xiang-ying, ZI Ling-shan, LU Qing-qing, LI Rong-chun. Cloning and Bioinformatics Analysis of the Ergothioneine Biosynthesis Genes in Naematelia aurantialba and Stereum hirsutum [J]. Biotechnology Bulletin, 2024, 40(7): 259-272. |
| [10] | HE Yu-bing, FU Zhen-hao, LI Ren-han, LIU Xiu-xia, LIU Chun-li, YANG Yan-kun, LI Ye, BAI Zhong-hu. Efficient Biosynthesis of 2-Naphthaleneethanol in Metabolically Engineered Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2024, 40(7): 99-107. |
| [11] | HU Jin-jin, LI Su-zhen, MA Xu-hui, LIU Xiao-qing, XIE Shan-shan, JIANG Hai-yang, CHEN Ru-mei. Regulation of Maize Anthocyanin Biosynthesis Metabolism [J]. Biotechnology Bulletin, 2024, 40(6): 34-44. |
| [12] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [13] | JI Hong-chao, LI Zheng-yan. Research Progress and Prospects in the Structural Annotation of Unknown Secondary Metabolites Based on Mass Spectrometry [J]. Biotechnology Bulletin, 2024, 40(10): 76-85. |
| [14] | WANG Jun-fang, HUANG Qiu-bin, ZHANG Piao-dan, ZHANG Peng-pai. Structure and Biosynthesis of Surfactin as well as Its Role in Biological Control [J]. Biotechnology Bulletin, 2024, 40(1): 100-112. |
| [15] | CHEN Zhi-min, LI Cui, WEI Ji-tian, LI Xin-ran, LIU Yi, GUO Qiang. Research Progress in the Regulation of Chlorogenic Acid Biosynthesis and Its Application [J]. Biotechnology Bulletin, 2024, 40(1): 57-71. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||