








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 44-53.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0222
Previous Articles Next Articles
WANG Fang( ), SHAO Hui-ru, LYU Lin-long, ZHAO Dian, HU Zhen, LYU Jian-zhen(
), SHAO Hui-ru, LYU Lin-long, ZHAO Dian, HU Zhen, LYU Jian-zhen( ), JIANG Liang(
), JIANG Liang( )
)
Received:2025-03-04
Online:2025-09-26
Published:2025-09-24
Contact:
LYU Jian-zhen, JIANG Liang
E-mail:2897447253@qq.com;lvjianzhen110@163.com;jiangliang188@gmail.com
WANG Fang, SHAO Hui-ru, LYU Lin-long, ZHAO Dian, HU Zhen, LYU Jian-zhen, JIANG Liang. Establishment of TurboID Proximity Labeling Technology in Plants and Bacteria[J]. Biotechnology Bulletin, 2025, 41(9): 44-53.
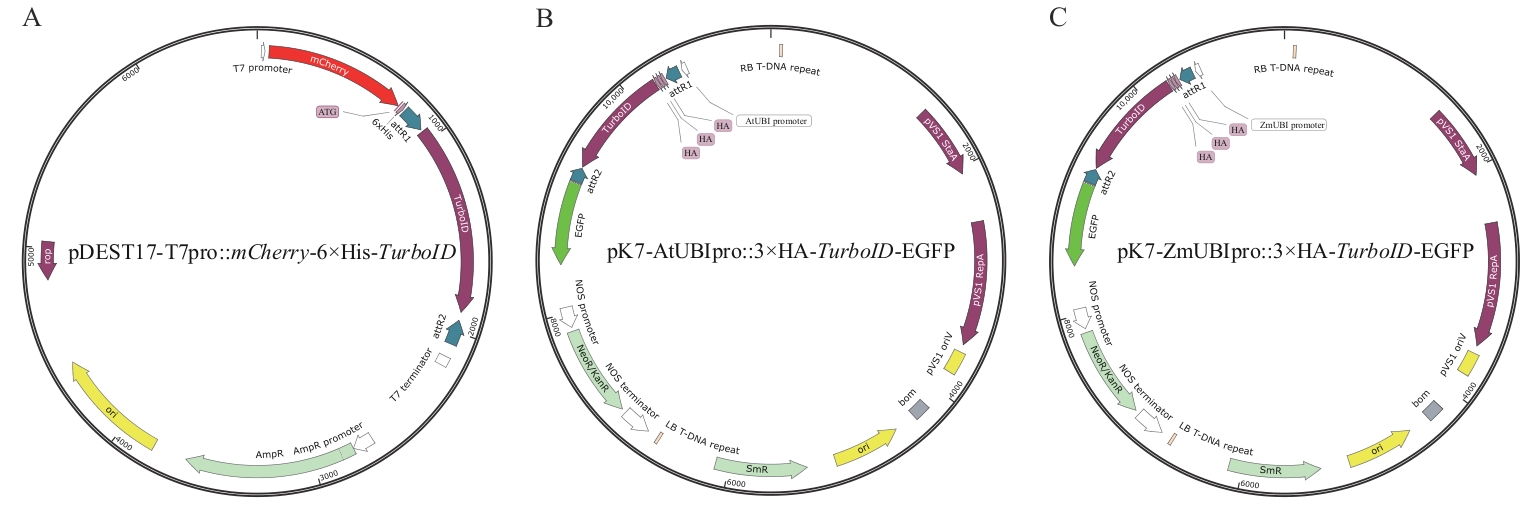
Fig. 1 Expression vectors for TurboID proximity labeling in plants and bacteriaA: Expression vector (pDEST17-T7pro::mCherry-6×His-TurboID) for TurboID proximity labeling in Escherichia coli. B: Expression vector (pK7-AtUBIpro::3×HA-TurboID-EGFP) for TurboID proximity labeling in dicotyledonous plants, including Arabidopsis thalian, Solanum lycopersicum, and Nicotiana benthamiana. C: Expression vector (pK7-ZmUBIpro::3×HA-TurboID-EGFP) for TurboID proximity labeling in monocotyledonous plant (Oryza sativa)
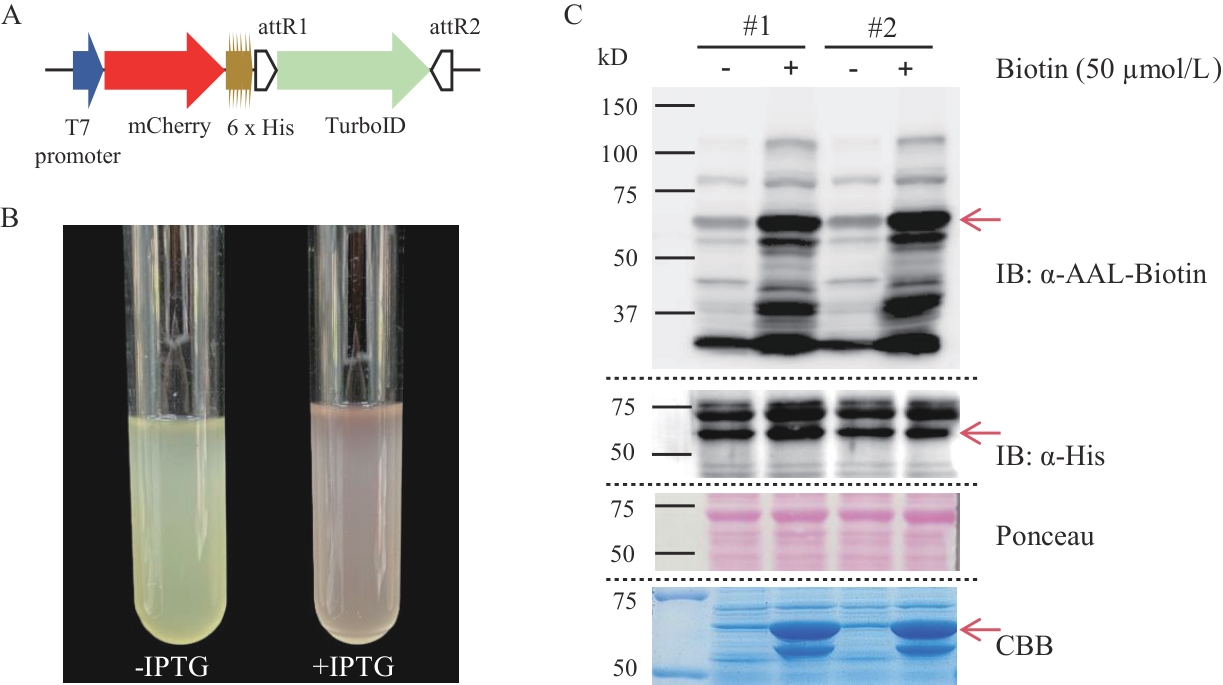
Fig. 3 Establishment of the TurboID proximity labeling method in E. coliA: Schematic diagram of the expression elements of the fusion protein mCherry-TurboID3. B: Photographs of bacterial cultures with and without IPTG induction. C: Western-blot detection of the expression of the fusion protein and biotinylation of proximity labeling in two transgenic E. coli strains (#1; #2). "-" and "+" indicate treatment with ddH2O and 50 μmol/L biotin solution respectively, in the Coomassie Brilliant Blue staining image, they indicate the uninduced and induced states respectively

Fig. 4 Optimization of biotin concentration and labeling time used in proximity labelingA: Treat the induced bacterial culture with biotin solutions of different concentrations for 3 h and then perform Western-blot detection using α-AAL-Biotin and α-His antibodies. B: Treat the induced bacterial culture with a 50 μmol/L biotin solution for different durations and then perform Western-blot detection

Fig. 5 Establishment of the TurboID proximity labeling method in ArabidopsisA: Schematic diagram of the expression elements of the fusion protein TurboID-EGFP. B: Phenotypic comparison of 30-day-old seedlings between wild-type and transgenic Arabidopsis thaliana. C: Western-blot detection of the expression of fusion proteins and biotinylation of proximity labeling in two transgenic Arabidopsis lines (#3; #5). "-" and "+" indicate treatment with ddH2O and 100 μmol/L biotin solution respectively. The same below

Fig. 6 Establishment of the TurboID proximity labeling method in S. lycopersicumA: Comparative analysis of 30-day-old wild-type and transgenic Solanum lycopersicum. B: Western-blot detection of the expression of fusion proteins and biotinylation of proximity labeling in the leaves of two transgenic tomato lines (#1; #2)

Fig.7 Establishment of the TurboID proximity labeling method based on tobacco transient expressionA: Image of tobacco leaf injection. Ruby-expressing Agrobacterium-injected tobacco serves as a visual control for tobacco expression, and CK being the leaf not injected with any Agrobacterium. B: Western-blot detection of the expression of fusion proteins and biotinylation of proximity labeling. From left to right, the three samples are tobacco leaves injected with Agrobacterium but not biotin, leaves not injected with any Agrobacterium or biotin, and tobacco leaves injected with both Agrobacterium and 20 μmol/L biotin
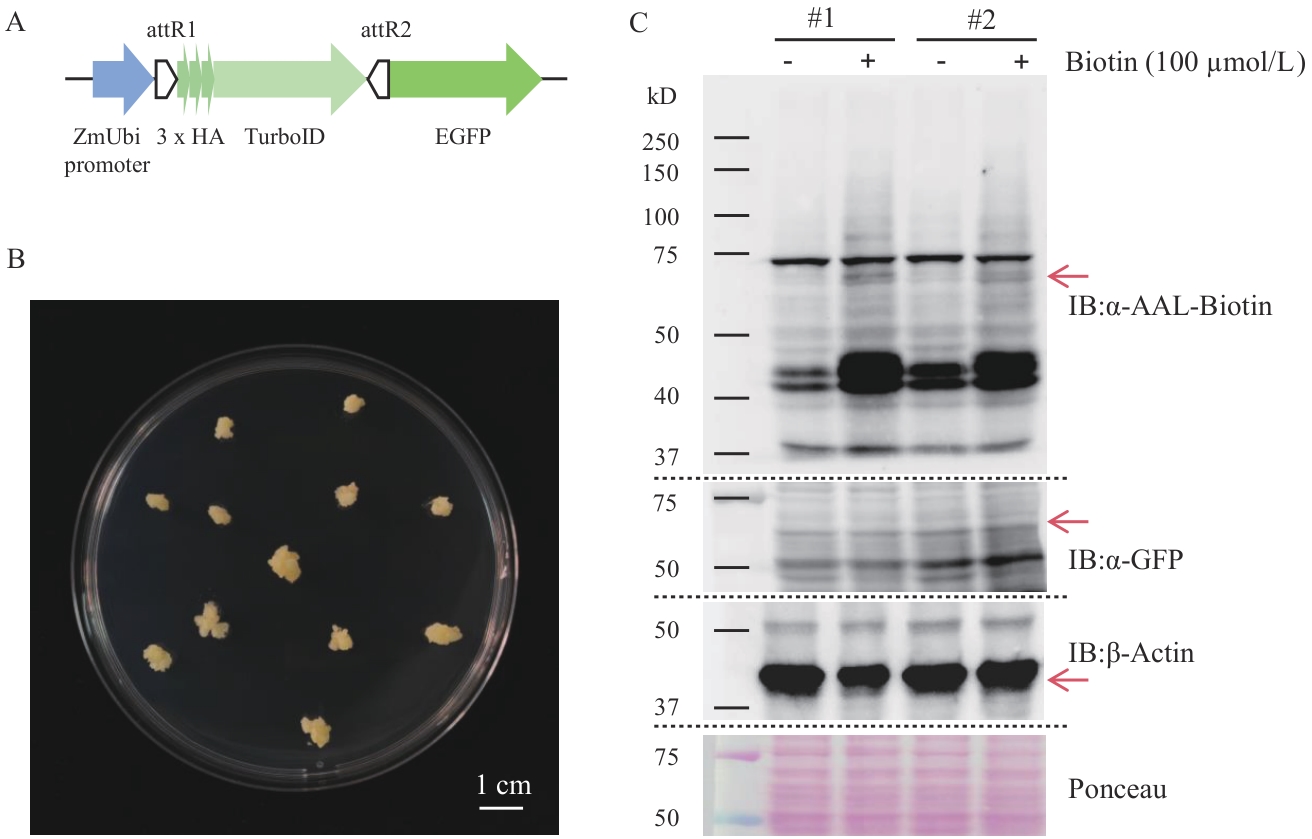
Fig. 8 Establishment of the TurboID proximity labeling method in rice callusesA: Schematic diagram of the expression elements of the fusion protein TurboID-EGFP. B: Image of rice calluses. C: Western-blot detection of the expression of the fusion protein and biotinylation of proximity labeling in two transgenic rice lines (#1; #2)
| [1] | Gingras AC, Abe KT, Raught B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles [J]. Curr Opin Chem Biol, 2019, 48: 44-54. |
| [2] | Duarte CEM, Euclydes NC. Protein-protein interaction via two-hybrid assay in yeast [J]. Methods Mol Biol, 2024, 2724: 193-210. |
| [3] | Lo SF. Co-immunoprecipitation (co-ip) in mammalian cells [J]. Methods Mol Biol, 2023, 2655: 67-77. |
| [4] | Zhang YJ, Natale R, Domingues Júnior AP, et al. Rapid identification of protein-protein interactions in plants [J]. Curr Protoc Plant Biol, 2019, 4(4): e20099. |
| [5] | Douzi B. Protein-protein interactions: surface plasmon resonance [J]. Methods Mol Biol, 2017, 1615: 257-275. |
| [6] | Roux KJ, Kim DI, Raida M, et al. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells [J]. J Cell Biol, 2012, 196(6): 801-810. |
| [7] | Choi CR, Rhee HW. Proximity labeling: an enzymatic tool for spatial biology [J]. Trends Biotechnol, 2022, 40(2): 145-148. |
| [8] | Sato S, Yoshida M, Hatano K, et al. N′-acyl-N-methylphenylenediamine as a novel proximity labeling agent for signal amplification in immunohistochemistry [J]. Bioorg Med Chem, 2019, 27(6): 1110-1118. |
| [9] | Rhee HW, Zou P, Udeshi ND, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging [J]. Science, 2013, 339(6125): 1328-1331. |
| [10] | Loh KH, Stawski PS, Draycott AS, et al. Proteomic analysis of unbounded cellular compartments: synaptic clefts [J]. Cell, 2016, 166(5): 1295-1307.e21. |
| [11] | Hopkins C, Gibson A, Stinchcombe J, et al. Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope [J]. Meth Enzymol, 2000, 327: 35-45. |
| [12] | Martell JD, Deerinck TJ, Sancak Y, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy [J]. Nat Biotechnol, 2012, 30(11): 1143-1148. |
| [13] | Trinkle-Mulcahy L. Recent advances in proximity-based labeling methods for interactome mapping [J]. F1000Res, 2019, 8(F1000 Faculty Rev): 135. |
| [14] | Parrott MB, Barry MA. Metabolic biotinylation of secreted and cell surface proteins from mammalian cells [J]. Biochem Biophys Res Commun, 2001, 281(4): 993-1000. |
| [15] | Jeong KH, Son SB, Ko JH, et al. Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor [J]. Biochem Biophys Res Commun, 2024, 733: 150601. |
| [16] | Kim DI, Roux KJ. Filling the void: proximity-based labeling of proteins in living cells [J]. Trends Cell Biol, 2016, 26(11): 804-817. |
| [17] | Kim DI, Jensen SC, Noble KA, et al. An improved smaller biotin ligase for BioID proximity labeling [J]. Mol Biol Cell, 2016, 27(8): 1188-1196. |
| [18] | Branon TC, Bosch JA, Sanchez AD, et al. Efficient proximity labeling in living cells and organisms with TurboID [J]. Nat Biotechnol, 2018, 36(9): 880-887. |
| [19] | Zhang YL, Song GY, Lal NK, et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity [J]. Nat Commun, 2019, 10(1): 3252. |
| [20] | Mair A, Xu SL, Branon TC, et al. Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID [J]. eLife, 2019, 8: e47864. |
| [21] | Arora D, Abel NB, Liu C, et al. Establishment of proximity-dependent biotinylation approaches in different plant model systems [J]. Plant Cell, 2020, 32(11): 3388-3407. |
| [22] | Kreis E, König K, Misir M, et al. TurboID reveals the proxiomes of Chlamydomonas proteins involved in thylakoid biogenesis and stress response [J]. Plant Physiol, 2023, 193(3): 1772-1796. |
| [23] | Sun FA, Hamada N, Montes C, et al. TurboID-based proteomic profiling reveals proxitome of ASK1 and CUL1 of the SCF ubiquitin ligase in plants [J]. New Phytol, 2024, 244(6): 2127-2136. |
| [24] | Li PP, Li JJ, Wang L, et al. Proximity labeling of interacting proteins: application of BioID as a discovery tool [J]. Proteomics, 2017, 17(20): 1700002. |
| [25] | Cho KF, Branon TC, Udeshi ND, et al. Proximity labeling in mammalian cells with TurboID and split-TurboID [J]. Nat Protoc, 2020, 15(12): 3971-3999. |
| [26] | 邝嘉怡, 李洪清, 沈文锦, 等. 基于TurboID的植物蛋白邻近标记实验方法 [J]. 植物学报, 2021, 56(5): 584-593. |
| Kuang JY, Li HQ, Shen WJ, et al. Methods for TurboID-based proximal labeling in plants [J]. Chin Bull Bot, 2021, 56(5): 584-593. | |
| [27] | 刘佳欣, 何明良, 刘颖湘, 等. 利用TurboID邻近蛋白标记技术获得水稻互作蛋白组的实验方法 [J]. 土壤与作物, 2023, 12(3): 256-263. |
| Liu JX, He ML, Liu YX, et al. Experimental method for obtaining interaction proteome using TurboID-based proximity labeling technology in rice [J]. Soils Crops, 2023, 12(3): 256-263. | |
| [28] | May DG, Scott KL, Campos AR, et al. Comparative application of BioID and TurboID for protein-proximity biotinylation [J]. Cells, 2020, 9(5): 1070. |
| [29] | Tan H, Zhou Y, Dinius E, et al. The Ti-TAN plasmid toolbox for TurboID-based proximity labeling assays in Nicotiana benthamiana [J]. J Integr Plant Biol, 2024, 66(2): 166-168. |
| [30] | Cho KF, Branon TC, Rajeev S, et al. Split-TurboID enables contact-dependent proximity labeling in cells [J]. Proc Natl Acad Sci USA, 2020, 117(22): 12143-12154. |
| [31] | Strotmann VI, Stahl Y. Visualization of in vivo protein-protein interactions in plants [J]. J Exp Bot, 2022, 73(12): 3866-3880. |
| [32] | Garloff V, Krüger T, Brakhage A, et al. Control of TurboID-dependent biotinylation intensity in proximity ligation screens [J]. J Proteom, 2023, 279: 104886. |
| [33] | Yang XX, Wen ZY, Zhang DL, et al. Proximity labeling: an emerging tool for probing in planta molecular interactions [J]. Plant Commun, 2020, 2(2): 100137. |
| [34] | Zhang YL, Li YY, Yang XX, et al. TurboID-based proximity labeling for in planta identification of protein-protein interaction networks [J]. J Vis Exp, 2020(159): 10.3791/60728. |
| [35] | Feng C, Roitinger E, Hudecz O, et al. TurboID-based proteomic profiling of meiotic chromosome axes in Arabidopsis thaliana [J]. Nat Plants, 2023, 9(4): 616-630. |
| [36] | Li XF, Wei YP, Fei QL, et al. TurboID-mediated proximity labeling for screening interacting proteins of FIP37 in Arabidopsis [J]. Plant Direct, 2023, 7(12): e555. |
| [37] | Xiong ZR, Lo HP, McMahon KA, et al. In vivo proteomic mapping through GFP-directed proximity-dependent biotin labelling in zebrafish [J]. eLife, 2021, 10: e64631. |
| [38] | Kim HB, Kim KE. Precision proteomics with TurboID: mapping the suborganelle landscape [J]. Korean J Physiol Pharmacol, 2024, 28(6): 495-501. |
| [39] | Merta H, Gov K, Isogai T, et al. Spatial proteomics of ER tubules reveals CLMN, an ER-actin tether at focal adhesions that promotes cell migration [J]. Cell Rep, 2025, 44(4): 115502. |
| [1] | LI Ya-qiong, GESANG La-mao, CHEN Qi-di, YANG Yu-huan, HE Hua-zhuan, ZHAO Yao-fei. Heterologous Overexpression of Sorghum SbSnRK2.1 Enhances the Resistance to Salt Stress in Arabidopsis [J]. Biotechnology Bulletin, 2025, 41(8): 115-123. |
| [2] | DENG Mei-bi, YAN Lang, ZHAN Zhi-tian, ZHU Min, HE Yu-bing. Efficient CRISPR Gene Editing in Rice Assisted by RUBY [J]. Biotechnology Bulletin, 2025, 41(8): 65-73. |
| [3] | WANG Cong-huan, WU Guo-qiang, WEI Ming. Functional Mechanism of Plant CBL in Regulating the Responses to Abiotic and Biotic Stresses [J]. Biotechnology Bulletin, 2025, 41(7): 1-16. |
| [4] | HUANG Dan-dan, WU Yun-yi, ZOU Jian-hua, YU Ting, ZHU Yan-hui, YANG Mei-hong, DONG Wen-li, GAO Dong-li. Cloning and Interaction Analysis of StPTST2a Gene in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 172-180. |
| [5] | LIU Tong-tong, LI Xiao-hui, YANG Jun-long, CHEN Wang, YU Meng, WANG Chao-fan, WANG Feng-ru, KE Shao-ying. Functional Study on ZmSTART1 Regulation of Maize Vascular Bundle Formation [J]. Biotechnology Bulletin, 2025, 41(4): 115-122. |
| [6] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [7] | ZHANG Yi-heng, LIU Jia-zheng, WANG Xue-chen, SUN Zheng-zhe, XUE Ya-jun, WANG Pei, HAN Hua, ZHENG Hong-wei, LI Xiao-juan. Dynamic Changes of Arabidopsis Endoplasmic Reticulum Based on Enhanced Super-resolution Images [J]. Biotechnology Bulletin, 2024, 40(4): 67-76. |
| [8] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [9] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [10] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [11] | LI Yi-jun, WU Chen-chen, LI Rui, WANG Zhe, HE Shan-wen, WEI Shan-jun, ZHANG Xiao-xia. Exploring Cultivation Approaches for New Endophytic Bacterial Resource in Oryza sativa [J]. Biotechnology Bulletin, 2023, 39(4): 201-211. |
| [12] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [13] | YANG Yi-shan, SUN Ping-yong, YU Mu-lan. QTL Mapping for Resistance to Rice Kernel Smut of Male Sterile Line [J]. Biotechnology Bulletin, 2022, 38(3): 16-21. |
| [14] | TANG Yue-hui, ZHAO Yu-fan, LIN Jin, WANG Yin, CAO Bo-yuan, CHE Yi-fan, YANG Wen-jie, BAO Xin-xin, YANG Tong-wen. Identification and Gene Mapping of a Seedling Lethal Mutant in Rice [J]. Biotechnology Bulletin, 2022, 38(10): 124-131. |
| [15] | YANG Hua-jie, ZHOU Yu-ping, FAN Tian, LV Tian-xiao, XIE Chu-ping, TIAN Chang-en. Screening and Identification of IQM4-Interacting Proteins in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2021, 37(11): 190-196. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||