








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 114-124.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0525
Previous Articles Next Articles
HE Qi-chen( ), YANG Yang, ALIYA Waili, TANG Xin-yue, LI Zhong-xi, CHEN Yong-kun, CHEN Ling-na(
), YANG Yang, ALIYA Waili, TANG Xin-yue, LI Zhong-xi, CHEN Yong-kun, CHEN Ling-na( )
)
Received:2025-05-22
Online:2026-01-26
Published:2026-02-04
Contact:
CHEN Ling-na
E-mail:3063801384@qq.com;ln.chen@xjnu.edu.cn
HE Qi-chen, YANG Yang, ALIYA Waili, TANG Xin-yue, LI Zhong-xi, CHEN Yong-kun, CHEN Ling-na. Investigation into the Family Characteristics of the Lavender Copper Amine Oxidase Gene and the Role of LaCuAO1 in Bioamine Degradation[J]. Biotechnology Bulletin, 2026, 42(1): 114-124.
在线生物学软件 Online biological software | 网站地址 Website address |
|---|---|
| Phytozome v13数据库 | https://phytozome-next.jgi.doe.gov/ |
| CDD | https://www.ncbi.nlm.nih.gov/cdd |
| HMMER | http://www.hmmer.org/ |
| GSDS | http://gsds.gao-lab.org/ |
| ExPASy | https://web.expasy.org/protparam/ |
| SOPMA | https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl page=npsa sopma |
| WoLFPSORT | http://wolfpsort.seq.cbrc.jp |
| iTOL | https://itol.embl.de/ |
| PlantCARE | http://bioinformatics.psb.ugent.be/wetools/plantcare/html/ |
Table 1 Relevant bioinformatics software and websites
在线生物学软件 Online biological software | 网站地址 Website address |
|---|---|
| Phytozome v13数据库 | https://phytozome-next.jgi.doe.gov/ |
| CDD | https://www.ncbi.nlm.nih.gov/cdd |
| HMMER | http://www.hmmer.org/ |
| GSDS | http://gsds.gao-lab.org/ |
| ExPASy | https://web.expasy.org/protparam/ |
| SOPMA | https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl page=npsa sopma |
| WoLFPSORT | http://wolfpsort.seq.cbrc.jp |
| iTOL | https://itol.embl.de/ |
| PlantCARE | http://bioinformatics.psb.ugent.be/wetools/plantcare/html/ |
基因名称 Gene ID | 引物序列 Primer sequence (5′‒3′) | 基因长度 Product length (bp) |
|---|---|---|
| La05G00442 | F: GGAGTGGACAGCTTACAGGC | 187 |
| R: CCTTCACCGACACGTGGATT | ||
| La05G01835 | F: GTGTAGGCGAAGGATTGGCT | 181 |
| R: GCCGGAGAGCAGTTGAAGAA | ||
| La25G00173 | F: GTCGGTCGCCAACTACGATT | 188 |
| R: CGTGGATAACACCGACGACA | ||
| La15G00866 | F: GGATCCTTCCGAGTTCCACG | 171 |
| R: TTGTACGGCGTCACCCAAAT | ||
| La00G03915 | F: CTCTCTGGAGCAACCCGAAG | 194 |
| R: GCACAAACAGCTCGGAAGTG | ||
| La10G00763 | F: AGGGCCTAGCTTTCGGATCA | 180 |
| R: TCTCCATAGGGCACGACCAT | ||
| LaActin | F:GGTCGTACAACTGGTATTGTTC R:TCCAGCAGCTTCCATTCCGATCA | 139 |
Table 2 RT-qPCR primer sequences
基因名称 Gene ID | 引物序列 Primer sequence (5′‒3′) | 基因长度 Product length (bp) |
|---|---|---|
| La05G00442 | F: GGAGTGGACAGCTTACAGGC | 187 |
| R: CCTTCACCGACACGTGGATT | ||
| La05G01835 | F: GTGTAGGCGAAGGATTGGCT | 181 |
| R: GCCGGAGAGCAGTTGAAGAA | ||
| La25G00173 | F: GTCGGTCGCCAACTACGATT | 188 |
| R: CGTGGATAACACCGACGACA | ||
| La15G00866 | F: GGATCCTTCCGAGTTCCACG | 171 |
| R: TTGTACGGCGTCACCCAAAT | ||
| La00G03915 | F: CTCTCTGGAGCAACCCGAAG | 194 |
| R: GCACAAACAGCTCGGAAGTG | ||
| La10G00763 | F: AGGGCCTAGCTTTCGGATCA | 180 |
| R: TCTCCATAGGGCACGACCAT | ||
| LaActin | F:GGTCGTACAACTGGTATTGTTC R:TCCAGCAGCTTCCATTCCGATCA | 139 |
基因ID号 Gene ID | 氨基酸数 Number of amino acids | 分子量 Molecular weight (Da) | 理论等电点 Theoretical isoelectric point | 不稳定系数 Stability coefficient | 脂溶指数 Lipid solubility index | 平均亲水值 Average hydrophilicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| La03G02650 | 784 | 86 774.78 | 6.35 | 42.95 | 79.69 | -0.311 | 叶绿体 |
| La05G00442 | 785 | 86 936.93 | 6.42 | 43.81 | 78.10 | -0.330 | 叶绿体 |
| La05G01835 | 785 | 87 387.54 | 6.55 | 43.60 | 77.49 | -0.340 | 过氧化物酶体 |
| La10G00763 | 707 | 78 669.53 | 7.64 | 43.69 | 80.82 | -0.346 | 叶绿体 |
| La03G03013 | 785 | 86 486.22 | 6.31 | 41.75 | 86.43 | -0.206 | 叶绿体 |
| La01G00448 | 665 | 75 822.56 | 5.46 | 41.92 | 81.29 | -0.356 | 叶绿体 |
| La22G00998 | 654 | 74 774.69 | 5.22 | 44.80 | 82.81 | -0.315 | 液泡 |
| La25G00172 | 714 | 80 710.59 | 5.73 | 36.17 | 83.94 | -0.350 | 叶绿体基质 |
| La00G03916 | 713 | 80 614.59 | 5.83 | 36.36 | 84.60 | -0.332 | 叶绿体基质 |
| La25G00173 | 726 | 81 549.49 | 5.77 | 42.39 | 80.66 | -0.325 | 液泡 |
| La15G00866 | 716 | 80 817.15 | 5.99 | 35.11 | 82.65 | -0.300 | 液泡 |
| La00G03915 | 726 | 81 556.54 | 5.74 | 42.11 | 80.66 | -0.322 | 液泡 |
| La25G00209 | 407 | 46 128.07 | 5.41 | 36.77 | 74.25 | -0.318 | 线粒体 |
| La00G03795 | 638 | 72 196.13 | 5.84 | 37.99 | 81.10 | -0.343 | 细胞质 |
| La07G01349 | 640 | 72 428.37 | 8.46 | 39.48 | 82.62 | -0.283 | 液泡 |
| La02G00655 | 528 | 59 545.29 | 5.18 | 39.84 | 76.17 | -0.412 | 线粒体 |
| La00G05435 | 638 | 72 288.29 | 8.52 | 40.93 | 83.03 | -0.280 | 液泡 |
| La17G01323 | 638 | 72 288.29 | 8.52 | 40.93 | 83.03 | -0.280 | 液泡 |
| La00G03531 | 352 | 39 783.90 | 5.59 | 32.69 | 75.82 | -0.400 | 细胞质 |
Table 3 Physicochemical properties of LaCuAO family members
基因ID号 Gene ID | 氨基酸数 Number of amino acids | 分子量 Molecular weight (Da) | 理论等电点 Theoretical isoelectric point | 不稳定系数 Stability coefficient | 脂溶指数 Lipid solubility index | 平均亲水值 Average hydrophilicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| La03G02650 | 784 | 86 774.78 | 6.35 | 42.95 | 79.69 | -0.311 | 叶绿体 |
| La05G00442 | 785 | 86 936.93 | 6.42 | 43.81 | 78.10 | -0.330 | 叶绿体 |
| La05G01835 | 785 | 87 387.54 | 6.55 | 43.60 | 77.49 | -0.340 | 过氧化物酶体 |
| La10G00763 | 707 | 78 669.53 | 7.64 | 43.69 | 80.82 | -0.346 | 叶绿体 |
| La03G03013 | 785 | 86 486.22 | 6.31 | 41.75 | 86.43 | -0.206 | 叶绿体 |
| La01G00448 | 665 | 75 822.56 | 5.46 | 41.92 | 81.29 | -0.356 | 叶绿体 |
| La22G00998 | 654 | 74 774.69 | 5.22 | 44.80 | 82.81 | -0.315 | 液泡 |
| La25G00172 | 714 | 80 710.59 | 5.73 | 36.17 | 83.94 | -0.350 | 叶绿体基质 |
| La00G03916 | 713 | 80 614.59 | 5.83 | 36.36 | 84.60 | -0.332 | 叶绿体基质 |
| La25G00173 | 726 | 81 549.49 | 5.77 | 42.39 | 80.66 | -0.325 | 液泡 |
| La15G00866 | 716 | 80 817.15 | 5.99 | 35.11 | 82.65 | -0.300 | 液泡 |
| La00G03915 | 726 | 81 556.54 | 5.74 | 42.11 | 80.66 | -0.322 | 液泡 |
| La25G00209 | 407 | 46 128.07 | 5.41 | 36.77 | 74.25 | -0.318 | 线粒体 |
| La00G03795 | 638 | 72 196.13 | 5.84 | 37.99 | 81.10 | -0.343 | 细胞质 |
| La07G01349 | 640 | 72 428.37 | 8.46 | 39.48 | 82.62 | -0.283 | 液泡 |
| La02G00655 | 528 | 59 545.29 | 5.18 | 39.84 | 76.17 | -0.412 | 线粒体 |
| La00G05435 | 638 | 72 288.29 | 8.52 | 40.93 | 83.03 | -0.280 | 液泡 |
| La17G01323 | 638 | 72 288.29 | 8.52 | 40.93 | 83.03 | -0.280 | 液泡 |
| La00G03531 | 352 | 39 783.90 | 5.59 | 32.69 | 75.82 | -0.400 | 细胞质 |
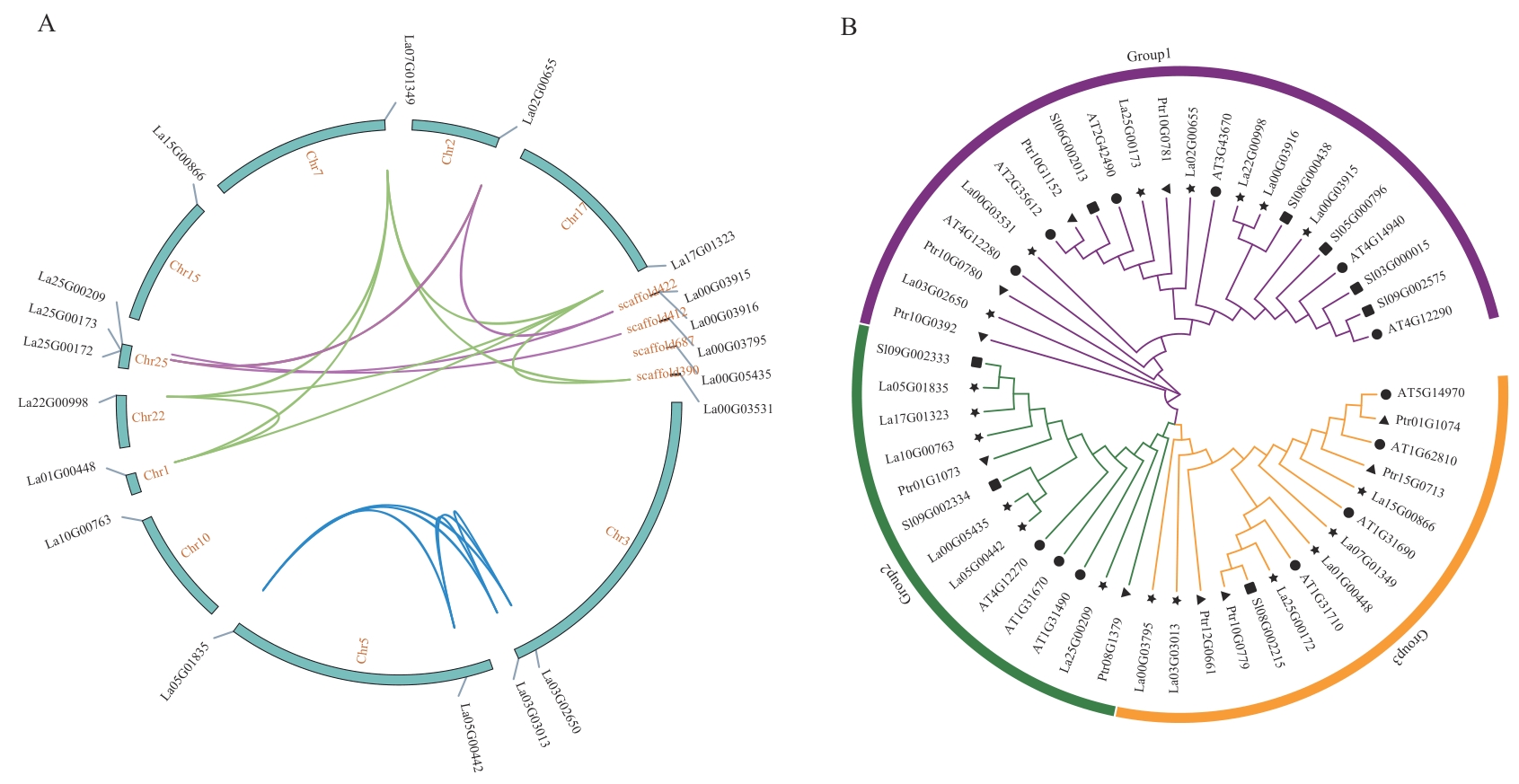
Fig. 3 Collinearity and phylogenetic analysis of LaCuAO family genes★ indicates Lavandula angustifolia CuAO. ▲ indicates Populus trichocarpa CuAO. ■ indicates Solanum lycopersicum CuAO. ● indicates Arabidopsis thaliana CuAO
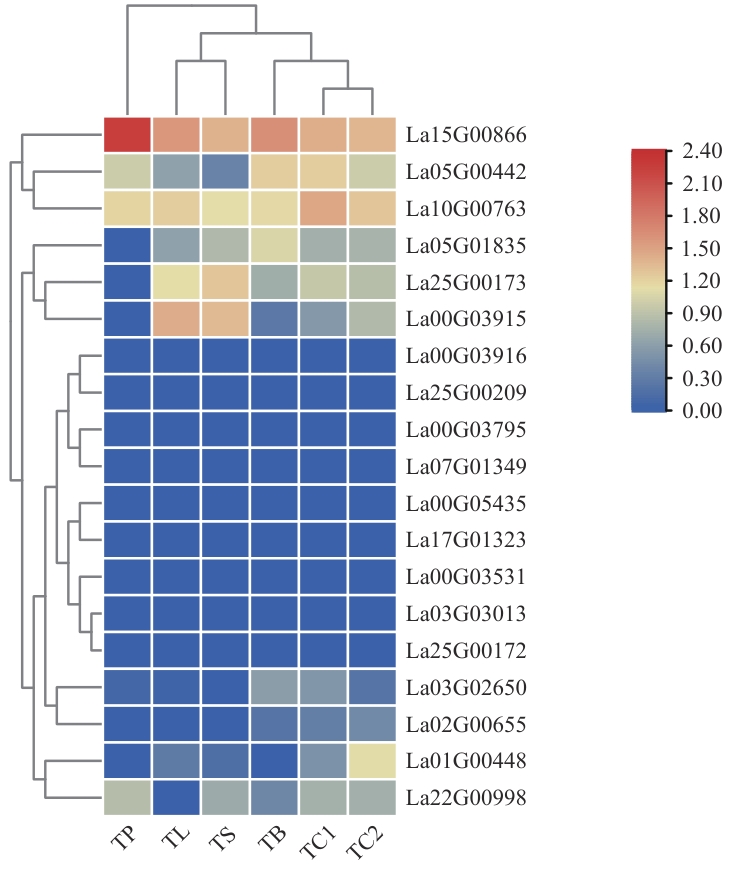
Fig. 5 Expressions of LaCuAO family genes in different tissues of lavenderTP: Petal. TL: Leaf. TS: Stem. TB: Flower Bud. TC1: Fresh calyx. TC2: Mature calyx
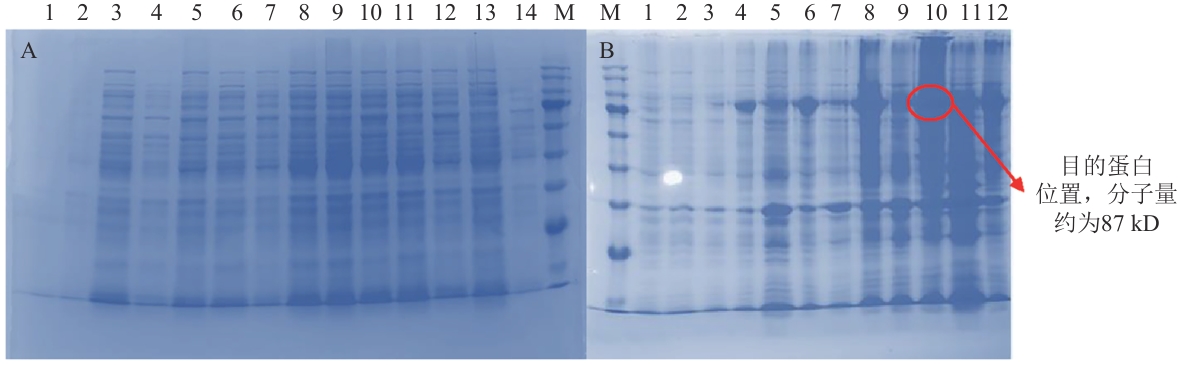
Fig. 7 SDS-PAGE gel electrophoresis patternA: Protein bands of the supernatant after centrifugation. B: Protein bands of precipitation. M: Marker. 1‒2: Blank control before and optimization (not induced). 3‒14 indicate the protein expressions sampled every 2 h at 2‒12 h after induction
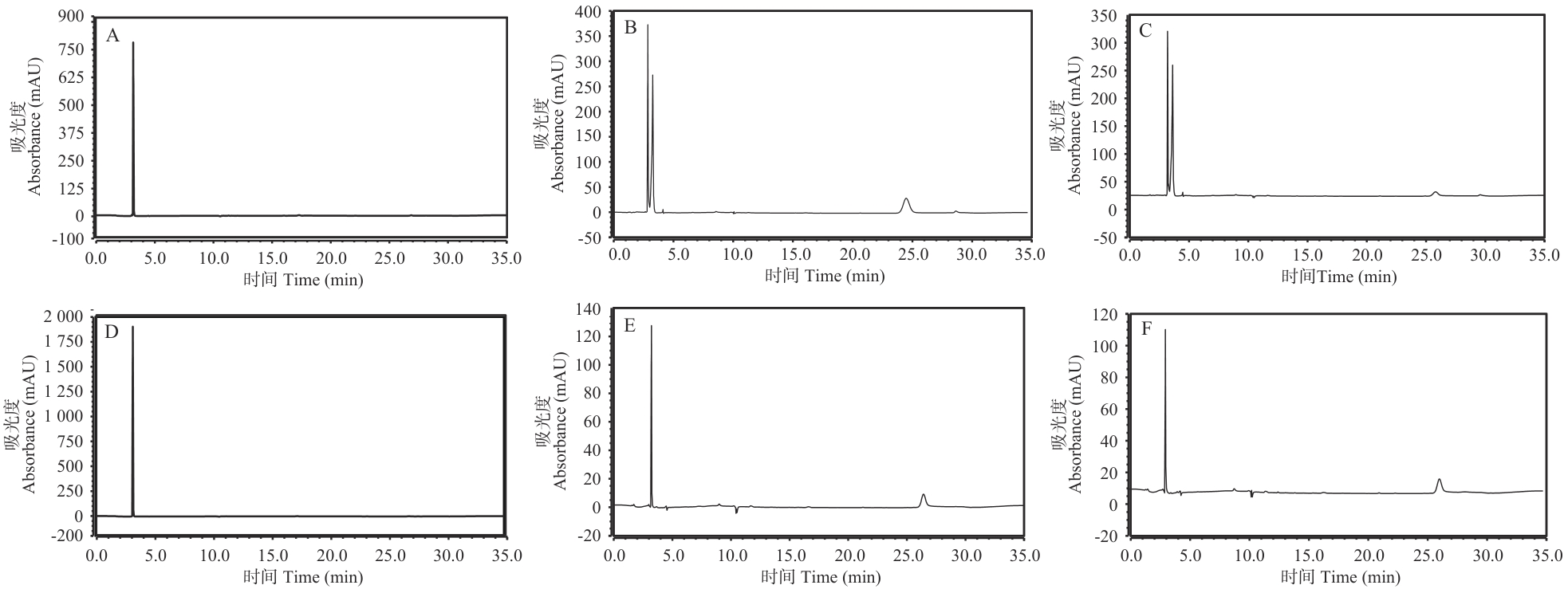
Fig. 8 HPLC degradation analysis of ethanolamine and histamine by optLaCuAO1A: Ethanolamine reference standards. B: Degradation of ethanolamine before codon optimization. C: Degradation of ethanolamine after codon optimization. D: Histamine reference standards. E: Degradation of histamine before codon optimization. F: Degradation of histamine after codon optimization
| [1] | Wang ZH, Wang Y, Yang J, et al. Reliable selection and holistic stability evaluation of reference genes for rice under 22 different experimental conditions [J]. Appl Biochem Biotechnol, 2016, 179(5): 753-775. |
| [2] | 屈海峰, 王登伟. 伊犁河谷油用薰衣草高产栽培技术 [J]. 新疆农垦科技, 2016, 39(8): 16-17. |
| Qu HF, Wang DW. High-yield cultivation techniques of oil lavender in Ili valley [J]. Xinjiang Farm Res Sci Technol, 2016, 39(8): 16-17. | |
| [3] | 陈国通, 左书瑞, 曹雪琴, 等. 吹扫捕集-气相色谱-质谱法测定薰衣草香气成分 [J]. 安徽农业科学, 2019, 47(2): 191-193. |
| Chen GT, Zuo SR, Cao XQ, et al. Determination of aroma components of lavender by purge and trap-gas chromatography-mass spectrometry [J]. J Anhui Agric Sci, 2019, 47(2): 191-193. | |
| [4] | Sharma M, Grewal K, Jandrotia R, et al. Essential oils as anticancer agents: potential role in malignancies, drug delivery mechanisms, and immune system enhancement [J]. Biomed Pharmacother, 2022, 146: 112514. |
| [5] | Crişan I, Ona A, Vârban D, et al. Current trends for lavender (Lavandula angustifolia Mill.) crops and products with emphasis on essential oil quality [J]. Plants, 2023, 12(2): 357. |
| [6] | Gonzalez ME, Jasso-Robles FI, Flores-Hernández E, et al. Current status and perspectives on the role of polyamines in plant immunity [J]. Ann Appl Biol, 2021, 178(2): 244-255. |
| [7] | 王伟, 程鑫, 崔振国, 等. 铜胺氧化酶基因PpCuAO4在桃成熟中的功能鉴定 [J]. 核农学报, 2022, 36(9): 1738-1745. |
| Wang W, Cheng X, Cui ZG, et al. Function identification of copper amine oxidase gene PpCuAO4 during ripening of peach (Prunus persica L.) fruit [J]. J Nucl Agric Sci, 2022, 36(9): 1738-1745. | |
| [8] | Yu TQ, Yin YF, Ge YH, et al. Enzymatic production of 4-hydroxyphenylacetaldehyde by oxidation of the amino group of tyramine with a recombinant primary amine oxidase [J]. Process Biochem, 2020, 92: 105-112. |
| [9] | Li JR, Wang YM, Dong YM, et al. The chromosome-based lavender genome provides new insights into Lamiaceae evolution and terpenoid biosynthesis [J]. Hortic Res, 2021, 8(1): 53. |
| [10] | Liu SP, Yao HL, Sun MF, et al. Heterologous expression and characterization of amine oxidases from Saccharopolyspora to reduce biogenic amines in huangjiu [J]. LWT, 2022, 169: 113963. |
| [11] | 徐圆圆, 赵国春, 郝颖颖, 等. 无患子RT-qPCR内参基因的筛选与验证 [J]. 生物技术通报, 2022, 38(10): 80-89. |
| Xu YY, Zhao GC, Hao YY, et al. Reference genes selection and validation for RT-qPCR in Sapindus mukorossi [J]. Biotechnol Bull, 2022, 38(10): 80-89. | |
| [12] | 岳俊齐, 约日耶提·萨力, 克拉热木·克里木江, 等. 薰衣草LaGGPPS5基因在大肠杆菌中催化萜类物质合成 [J]. 中国农业科技导报, 2023, 25(12): 85-92. |
| Yue JQ, Yueriyeti S, Kelaremu K, et al. Lavender LaGGPPS5 gene catalyzing terpenoids synthesis in Escherichia coli [J]. J Agric Sci Technol, 2023, 25(12): 85-92. | |
| [13] | 陈志军, 张立坚, 池青, 等. 小麦NAC转录因子TaNAC14的原核表达和分离纯化 [J]. 生物工程学报, 2024, 40(11): 4171-4182. |
| Chen ZJ, Zhang LJ, Chi Q, et al. Prokaryotic expression and purification of the transcription factor TaNAC14 in wheat (Triticum aestivum) [J]. Chin J Biotechnol, 2024, 40(11): 4171-4182. | |
| [14] | 姜维. 一株耐盐性高效生物胺降解新菌的筛选、分类鉴定及应用研究 [D]. 青岛: 中国海洋大学, 2014. |
| Jiang W. Study on the screening, taxonomic analysis and application of one novel, salt-tolerant and high efficient biogenic amines degrading bacterium [D]. Qingdao: Ocean University of China, 2014. | |
| [15] | 胡昌勤, 张夏, 常祎卓. HPLC分析的全生命周期管理 [J]. 中国现代应用药学, 2024, 41(20): 2758-2767. |
| Hu CQ, Zhang X, Chang YZ. Life-cycle management of HPLC analysis [J]. Chin J Mod Appl Pharm, 2024, 41(20): 2758-2767. | |
| [16] | 克拉热木·克里木江, 陈永坤, 李翠翠, 等. 薰衣草DXS基因家族的全基因组鉴定和表达分析 [J]. 基因组学与应用生物学, 2023, 42(9): 919-926. |
| Kelaremu·K, Chen YK, Li CC, et al. Genome-wide identification and expression analysis of DXS gene family in Lavandula angustifolia [J]. Genom Appl Biol, 2023, 42(9): 919-926. | |
| [17] | Kelimujiang K, Zhang WY, Zhang XX, et al. Genome-wide investigation of WRKY gene family in Lavandula angustifolia and potential role of LaWRKY57 and LaWRKY75 in the regulation of terpenoid biosynthesis [J]. Front Plant Sci, 2024, 15: 1449299. |
| [18] | Zhang ZB, Xiong T, Li KJ, et al. The tree of life of copper-containing amine oxidases [J]. Front Plant Sci, 2025, 16: 1544527. |
| [19] | Jakobsson E, Nilsson J, Ogg D, et al. Structure of human semicarbazide-sensitive amine oxidase/vascular adhesion protein-1 [J]. Acta Crystallogr D Biol Crystallogr, 2005, 61(Pt 11): 1550-1562. |
| [20] | 王鹏, 罗朝鹏, 盖江涛. 茄科植物甲基腐胺氧化酶基因进化路径分析 [J]. 分子植物育种, 2018, 16(1): 47-53. |
| Wang P, Luo ZP, Gai JT. Evolutionary path analysis of Methylputrescine oxidase gene in Solanaceae [J]. Mol Plant Breed, 2018, 16(1): 47-53. | |
| [21] | Malli RPN, Adal AM, Sarker LS, et al. De novo sequencing of the Lavandula angustifolia genome reveals highly duplicated and optimized features for essential oil production [J]. Planta, 2019, 249(1): 251-256. |
| [22] | Qu YN, Wang Q, Guo JH, et al. Peroxisomal CuAOζ and its product H2O2 regulate the distribution of auxin and IBA-dependent lateral root development in Arabidopsis [J]. J Exp Bot, 2017, 68(17): 4851-4867. |
| [23] | Zhang YN, Wang D, Li H, et al. Formation mechanism of glandular trichomes involved in the synthesis and storage of terpenoids in lavender [J]. BMC Plant Biol, 2023, 23(1): 307. |
| [24] | 张夏夏, 陈凌娜, 杨扬, 等. 薰衣草非特异性脂质转移蛋白基因nsLTP2-1和nsLTP2-2的克隆与功能分析 [J]. 植物遗传资源学报, 2024, 25(5): 834-843. |
| Zhang XX, Chen LN, Yang Y, et al. Cloning and functional analysis of nonspecific lipid transfer protein genes nsLTP2-1 and nsLTP2-2 in lavender [J]. J Plant Genet Resour, 2024, 25(5): 834-843. | |
| [25] | Meng FL, Zhao HN, Zhu B, et al. Genomic editing of intronic enhancers unveils their role in fine-tuning tissue-specific gene expression in Arabidopsis thaliana [J]. Plant Cell, 2021, 33(6): 1997-2014. |
| [26] | Wang J, Jiang WL, Yang ZH, et al. Cold-related genes BcCOR15B and BcCOR15A have environment adapted expression pattern in non-heading Chinese cabbage [J]. Plant Physiol Biochem, 2025, 221: 109567. |
| [27] | 秦善, 石洁, 潘徐盈, 等. 植物乳杆菌源胺氧化酶的异源表达及功能结构分析 [J]. 微生物学报, 2023, 63(12): 4698-4713. |
| Qin S, Shi J, Pan XY, et al. Heterologous expression, function and structure of amine oxidase derived from Lactiplantibacillus plantarum [J]. Acta Microbiol Sin, 2023, 63(12): 4698-4713. | |
| [28] | 倪秀梅, 方芳. 融合表达过氧化氢酶提高多铜氧化酶稳定性及降解生物胺能力 [J]. 生物工程学报, 2021, 37(12): 4382-4394. |
| Ni XM, Fang F. Fusion expression with catalase improves the stability of multicopper oxidase and its efficiency in degrading biogenic amines [J]. Chin J Biotechnol, 2021, 37(12): 4382-4394. | |
| [29] | 张阿娜, 韩雪, 谷天一, 等. 利用新型红酵母苯丙氨酸解氨酶制备低苯丙氨酸酪蛋白 [J]. 生物技术通报, 2024, 40(8): 309-319. |
| Zhang AN, Han X, Gu TY, et al. Preparation of low-phenylalanine casein by novel phenylalanine ammonia-lyases derived from Rhodotorula [J]. Biotechnol Bull, 2024, 40(8): 309-319. | |
| [30] | 张婵, 吴友根, 于靖, 等. 光与茉莉酸信号介导的萜类化合物合成分子机制 [J]. 生物技术通报, 2022, 38(8): 32-40. |
| Zhang C, Wu YG, Yu J, et al. Molecular mechanism of terpenoids synthesis intermediated by light and jasmonates signals [J]. Biotechnol Bull, 2022, 38(8): 32-40. | |
| [31] | Largeron M. Amine oxidases of the quinoproteins family: their implication in the metabolic oxidation of xenobiotics [J]. Ann Pharm Françaises, 2011, 69(1): 53-61. |
| [1] | WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought [J]. Biotechnology Bulletin, 2026, 42(1): 198-207. |
| [2] | GUO Tao, AI Li-jiao, ZOU Shi-hui, ZHOU Ling, LI Xue-mei. Functional Study of CjRAV1 from Camellia japonica in Regulating Flowering Delay [J]. Biotechnology Bulletin, 2025, 41(6): 208-217. |
| [3] | ZHOU Yi, LIU Yong-bo. Research Progress in the Mechanisms and Functions of Gene Loss in Genome Evolution [J]. Biotechnology Bulletin, 2025, 41(6): 38-48. |
| [4] | LIU Yuan, ZHAO Ran, LU Zhen-fang, LI Rui-li. Research Progress in the Biological Metabolic Pathway and Functions of Plant Carotenoids [J]. Biotechnology Bulletin, 2025, 41(5): 23-31. |
| [5] | WANG Ting, WANG Yi-dan, REN Shu-jin, MA Ting, JIN Meng-jun, YANG Cheng-de. Cloning and Expression Analysis of Pectate Lyase Gene CcPL20552 from Colletotrichum coccodes [J]. Biotechnology Bulletin, 2025, 41(5): 290-299. |
| [6] | LIU Hong-li, MA Yi-dan, WANG Wan-ru, YANG Ya-ru, HE Dan, LIU Yi-ping, KONG De-zheng. Cloning and Functional Analysis of NnCYP707A1 Gene from Lotus [J]. Biotechnology Bulletin, 2025, 41(5): 197-207. |
| [7] | TIAN Qin, LIU Kui, WU Xiang-wei, JI Yuan-yuan, CAO Yi-bo, ZHANG Ling-yun. Functional Study of Transcription Factor VcMYB17 in Regulating Drought Tolerance in Blueberry [J]. Biotechnology Bulletin, 2025, 41(4): 198-210. |
| [8] | XIANG Chun-fan, LI Le-song, WANG Juan, LIANG Yan-li, YANG Sheng-chao, LI Meng-fei, ZHAO Yan. Functional Identification and Expression Analysis of Cinnamonyl Alcohol Dehydrogenase AsCAD in Angelica sinensis [J]. Biotechnology Bulletin, 2025, 41(2): 295-308. |
| [9] | LI Ming, LIU Xiang-yu, WANG Yi-na, HE Si-mei, SHA Ben-cai. Cloning and Functional Characterization of 6-OMT Gene Related to Isocorydine Biosynthesis in Dactylicapnos scandens [J]. Biotechnology Bulletin, 2025, 41(2): 309-320. |
| [10] | YANG Chen-xin, LI Meng-xiu, JIANG Tang, ZHANG Wen-e, PAN Xue-jun. Identification and Expression Analysis of Anthocyanin-associated R2R3-MYB Genes in Canna indica [J]. Biotechnology Bulletin, 2025, 41(12): 201-213. |
| [11] | MIAO Hao-xiang, ZHANG Ying, GUO Shi-peng, ZHANG Jian. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yielding γ-aminobutyric Acid-producing Lactobacillus brevis TCCC13007 [J]. Biotechnology Bulletin, 2025, 41(11): 166-176. |
| [12] | LIU Zi-qi, ZHONG Pei, LI Qin, GUO Cheng, ZHANG Yan-mei, ZHANG Nai-feng, TU Yan, DIAO Qi-yu, BI Yan-liang. Application and Progress of CRISPR/Cas9 Technology in Probiotic Editing [J]. Biotechnology Bulletin, 2025, 41(11): 89-99. |
| [13] | WU Ding-jie, CHEN Ying-ying, XU Jing, LIU Yuan, ZHANG Hang, LI Rui-li. Research Progress in Plant Gibberellin Oxidase and Its Functions [J]. Biotechnology Bulletin, 2024, 40(7): 43-54. |
| [14] | SUN Hui-qiong, ZHANG Chun-lai, WANG Xi-liang, XU Hong-shen, DOU Miao-miao, YANG Bo-hui, CHAI Wen-ting, ZHAO Shan-shan, JIANG Xiao-dong. Identification, Expression and DNA Variation Analysis of FLS Gene Family in Chenopodium quinoa [J]. Biotechnology Bulletin, 2024, 40(7): 172-182. |
| [15] | ZHANG Na, LIU Meng-nan, QU Zhan-fan, CUI Yi-ping, NI Jia-yao, WANG Hua-zhong. Alternative Translation of Wheat Enolase-encoding Gene ENO2 and Its Prokaryotic Expression [J]. Biotechnology Bulletin, 2024, 40(5): 112-119. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||