








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 42-50.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0608
Previous Articles Next Articles
LI Hui-hui( ), WANG Shu-jie, KANG Xiao-long(
), WANG Shu-jie, KANG Xiao-long( )
)
Received:2025-06-11
Online:2026-01-26
Published:2026-02-04
Contact:
KANG Xiao-long
E-mail:lihuihuitx@126.com;kangxl9527@126.com
LI Hui-hui, WANG Shu-jie, KANG Xiao-long. The Role of RNA-specific Adenosine Deaminase 1 in Innate Immunity and Inflammation[J]. Biotechnology Bulletin, 2026, 42(1): 42-50.
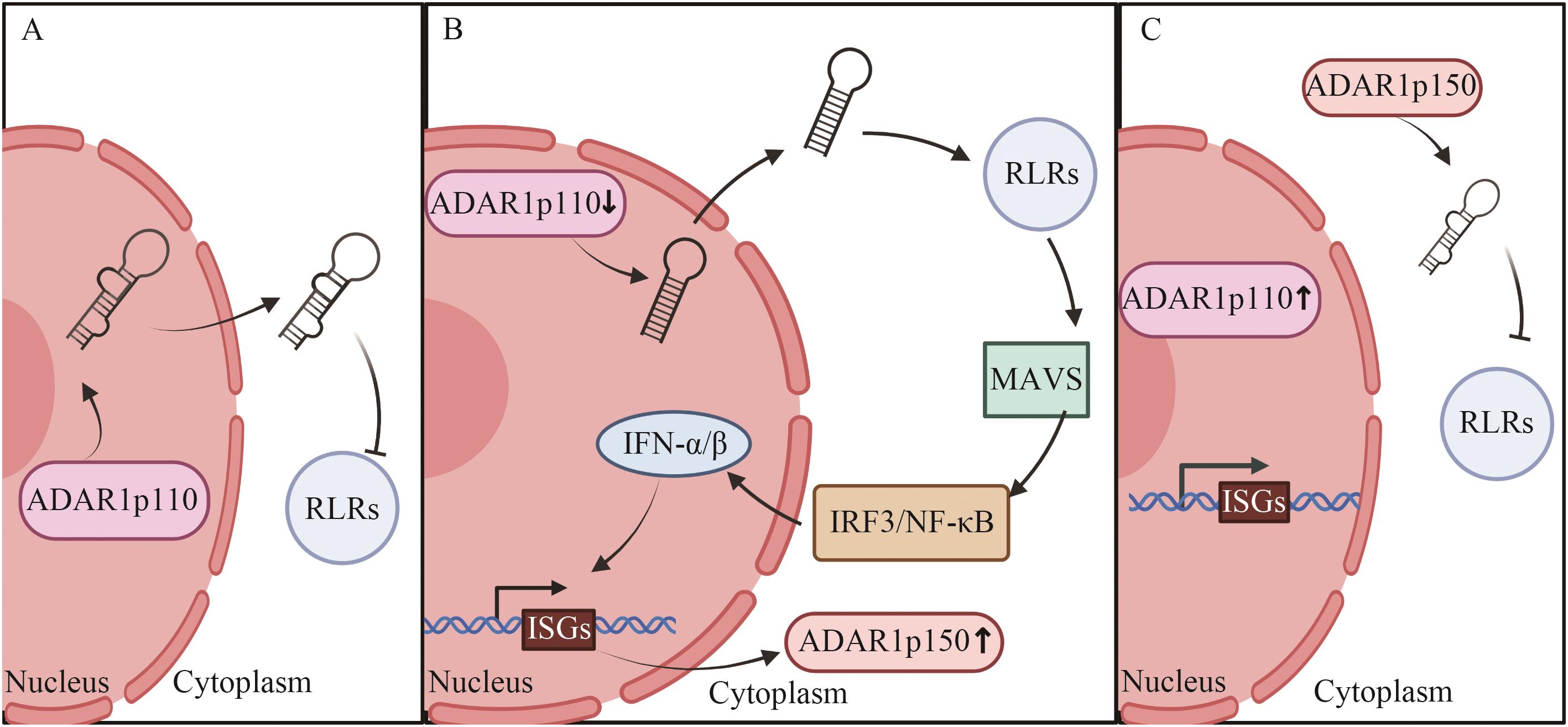
Fig. 1 Expression and function of two subtypes of ADAR1 in virus infectionA: When the virus is not infected. B: The early stage of viral infection. C: The late stage of viral infection. IFN-α/β: Type I IFN
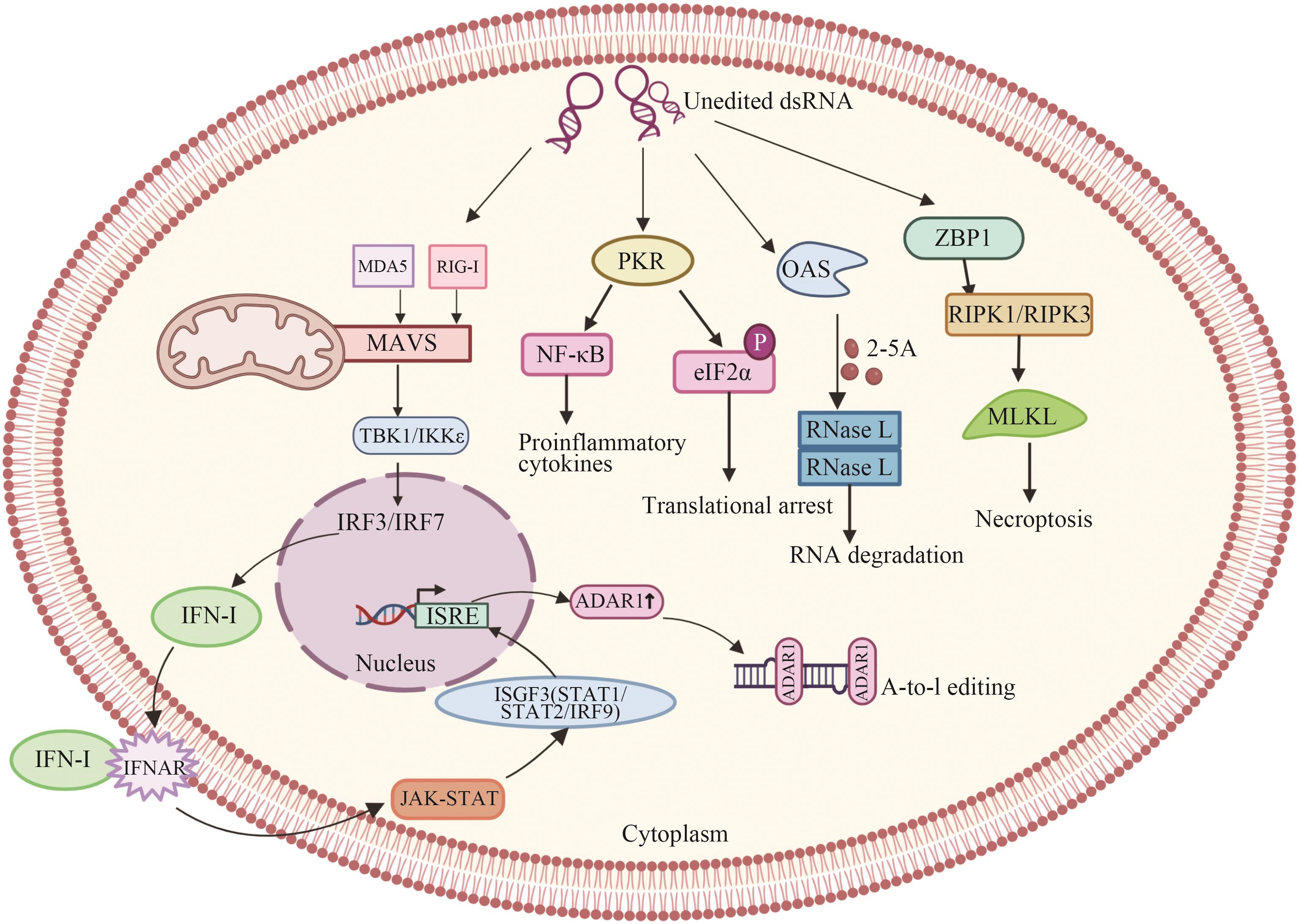
Fig. 2 Role of ADAR1 in innate immunityIRF3/IRF7: Transcription factor; NF-κB: nuclear factor-κB; IFN-Ⅰ: type I IFN; IFNAR: receptor for IFN; ISGs, interferon stimulating gene; ISGF3: interferon-stimulated gene factor 3 (ISGF3) is a transcription factor complex composed of STAT1, STAT2, and IRF9
| [1] | Song YL, Yang WB, Fu Q, et al. irCLASH reveals RNA substrates recognized by human ADARs [J]. Nat Struct Mol Biol, 2020, 27(4): 351-362. |
| [2] | Zhong YH, Zhong X, Qiao LJ, et al. Zα domain proteins mediate the immune response [J]. Front Immunol, 2023, 14: 1241694. |
| [3] | Bahn JH, Ahn J, Lin XZ, et al. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways [J]. Nat Commun, 2015, 6: 6355. |
| [4] | Song B, Shiromoto Y, Minakuchi M, et al. The role of RNA editing enzyme ADAR1 in human disease [J]. Wires RNA, 2022, 13(1): e1665. |
| [5] | Kleinova R, Rajendra V, Leuchtenberger AF, et al. The ADAR1 editome reveals drivers of editing-specificity for ADAR1-isoforms [J]. Nucleic Acids Res, 2023, 51(9): 4191-4207. |
| [6] | Ashley CN, Broni E, Miller WA. ADAR family proteins: a structural review [J]. Curr Issues Mol Biol, 2024, 46(5): 3919-3945. |
| [7] | Matthews MM, Thomas JM, Zheng YX, et al. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity [J]. Nat Struct Mol Biol, 2016, 23(5): 426-433. |
| [8] | Fisher AJ, Beal PA. Structural perspectives on adenosine to inosine RNA editing by ADARs [J]. Mol Ther Nucleic Acids, 2024, 35(3): 102284. |
| [9] | Quin J, Sedmík J, Vukić D, et al. ADAR RNA modifications, the epitranscriptome and innate immunity [J]. Trends Biochem Sci, 2021, 46(9): 758-771. |
| [10] | Song CZ, Sakurai M, Shiromoto Y, et al. Functions of the RNA editing enzyme ADAR1 and their relevance to human diseases [J]. Genes, 2016, 7(12): 129. |
| [11] | Xu L-D, Öhman M. ADAR1 editing and its role in cancer [J]. Genes, 2019, 10(1): 12. |
| [12] | Liddicoat BJ, Piskol R, Chalk AM, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself [J]. Science, 2015, 349(6252): 1115-1120. |
| [13] | Samuel CE. Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses [J]. J Biol Chem, 2019, 294(5): 1710-1720. |
| [14] | Yuan J, Xu L, Bao HJ, et al. Biological roles of A-to-I editing: implications in innate immunity, cell death, and cancer immunotherapy [J]. J Exp Clin Cancer Res, 2023, 42(1): 149. |
| [15] | Adamczak D, Fornalik M, Małkiewicz A, et al. ADAR1 expression in different cancer cell lines and its change under heat shock [J]. J Appl Genet, 2024: 149. |
| [16] | Szymczak F, Cohen-Fultheim R, Thomaidou S, et al. ADAR1-dependent editing regulates human β cell transcriptome diversity during inflammation [J]. Front Endocrinol, 2022, 13: 1058345. |
| [17] | Valentine A, Bosart K, Bush W, et al. Identification and characterization of ADAR1 mutations and changes in gene expression in human cancers [J]. Cancer Genet, 2024, 288/289: 82-91. |
| [18] | Mboukou A, Rajendra V, Messmer S, et al. Dimerization of ADAR1 modulates site-specificity of RNA editing [J]. Nat Commun, 2024, 15: 10051. |
| [19] | Roth SH, Danan-Gotthold M, Ben-Izhak M, et al. Increased RNA editing may provide a source for autoantigens in systemic lupus erythematosus [J]. Cell Rep, 2018, 23(1): 50-57. |
| [20] | Nakahama T, Kawahara Y. The RNA-editing enzyme ADAR1: a regulatory hub that tunes multiple dsRNA-sensing pathways [J]. Int Immunol, 2023, 35(3): 123-133. |
| [21] | Bazak L, Haviv A, Barak M, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes [J]. Genome Res, 2014, 24(3): 365-376. |
| [22] | Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs [J]. Nat Rev Mol Cell Biol, 2016, 17(2): 83-96. |
| [23] | Mu JQ, Wu C, Xu KM, et al. Conformational reorganization and phase separation drive hyper-editing of ADR-2-ADBP-1 complex [J]. Nucleic Acids Res, 2025, 53(5): gkaf148. |
| [24] | Uzonyi A, Nir R, Shliefer O, et al. Deciphering the principles of the RNA editing code via large-scale systematic probing [J]. Mol Cell, 2021, 81(11): 2374-2387.e3. |
| [25] | Rehwinkel J, Mehdipour P. ADAR1: from basic mechanisms to inhibitors [J]. Trends Cell Biol, 2025, 35(1): 59-73. |
| [26] | Deffit SN, Hundley HA. To edit or not to edit: regulation of ADAR editing specificity and efficiency [J]. Wiley Interdiscip Rev RNA, 2016, 7(1): 113-127. |
| [27] | Sharma M, Wagh P, Shinde T, et al. Exploring the role of pattern recognition receptors as immunostimulatory molecules [J]. Immunity Inflam & Disease, 2025, 13(5): e70150. |
| [28] | Chen RC, Zou J, Chen JW, et al. Pattern recognition receptors: function, regulation and therapeutic potential [J]. Signal Transduct Target Ther, 2025, 10(1): 216. |
| [29] | Lamers MM, van den Hoogen BG, Haagmans BL. ADAR1: “editor-in-chief” of cytoplasmic innate immunity [J]. Front Immunol, 2019, 10: 1763. |
| [30] | Sikorska J, Wyss DF. Recent developments in understanding RIG-I’s activation and oligomerization [J]. Sci Prog, 2024, 107(3): 368504241265182. |
| [31] | Liu SQ, Cai X, Wu JX, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation [J]. Science, 2015, 347(6227): aaa2630. |
| [32] | Ivashkiv LB, Donlin LT. Regulation of type I interferon responses [J]. Nat Rev Immunol, 2014, 14(1): 36-49. |
| [33] | Sampaio NG, Chauveau L, Hertzog J, et al. The RNA sensor MDA5 detects SARS-CoV-2 infection [J]. Sci Rep, 2021, 11(1): 13638. |
| [34] | Yang SY, Deng P, Zhu ZW, et al. Adenosine deaminase acting on RNA 1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs [J]. J Immunol, 2014, 193(7): 3436-3445. |
| [35] | Jiang YF, Zhang HY, Wang J, et al. Exploiting RIG-I-like receptor pathway for cancer immunotherapy [J]. J Hematol Oncol, 2023, 16(1): 8. |
| [36] | Dempsey LA. ADAR1 regulates mda5-MAVS [J]. Nat Immunol, 2016, 17(1): 47. |
| [37] | Shiromoto Y, Sakurai M, Minakuchi M, et al. ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells [J]. Nat Commun, 2021, 12(1): 1654. |
| [38] | Kim JI, Nakahama T, Yamasaki R, et al. RNA editing at a limited number of sites is sufficient to prevent MDA5 activation in the mouse brain [J]. PLoS Genet, 2021, 17(5): e1009516. |
| [39] | Deng P, Khan A, Jacobson D, et al. Adar RNA editing-dependent and-independent effects are required for brain and innate immune functions in Drosophila [J]. Nat Commun, 2020, 11(1): 1580. |
| [40] | Gal-Ben-Ari S, Barrera I, Ehrlich M, et al. PKR: a kinase to remember [J]. Front Mol Neurosci, 2019, 11: 480. |
| [41] | Hu SB, Heraud-Farlow J, Sun T, et al. ADAR1p150 prevents MDA5 and PKR activation via distinct mechanisms to avert fatal autoinflammation [J]. bioRxiv, 2023: 2023.01.25.525475. |
| [42] | Gannon HS, Zou T, Kiessling MK, et al. Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells [J]. Nat Commun, 2018, 9(1): 5450. |
| [43] | Pestal K, Funk CC, Snyder JM, et al. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development [J]. Immunity, 2015, 43(5): 933-944. |
| [44] | Sinigaglia K, Cherian A, Du QP, et al. An ADAR1 dsRBD3-PKR kinase domain interaction on dsRNA inhibits PKR activation [J]. Cell Rep, 2024, 43(8): 114618. |
| [45] | Clerzius G, Gélinas JF, Daher A, et al. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication [J]. J Virol, 2009, 83(19): 10119-10128. |
| [46] | Toth AM, Li ZQ, Cattaneo R, et al. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR [J]. J Biol Chem, 2009, 284(43): 29350-29356. |
| [47] | Gélinas JF, Clerzius G, Shaw E, et al. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase [J]. J Virol, 2011, 85(17): 8460-8466. |
| [48] | Wang YB, Holleufer A, Gad HH, et al. Length dependent activation of OAS proteins by dsRNA [J]. Cytokine, 2020, 126: 154867. |
| [49] | Daou S, Talukdar M, Tang JL, et al. A phenolic small molecule inhibitor of RNase L prevents cell death from ADAR1 deficiency [J]. Proc Natl Acad Sci USA, 2020, 117(40): 24802-24812. |
| [50] | Li YZ, Banerjee S, Goldstein SA, et al. Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line [J]. eLife, 2017, 6: e25687. |
| [51] | Jiao HP, Wachsmuth L, Kumari S, et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation [J]. Nature, 2020, 580(7803): 391-395. |
| [52] | Devos M, Tanghe G, Gilbert B, et al. Sensing of endogenous nucleic acids by ZBP1 induces keratinocyte necroptosis and skin inflammation [J]. J Exp Med, 2020, 217(7): e20191913. |
| [53] | Kesavardhana S, Subbarao Malireddi RK, Burton AR, et al. The Zα2 domain of ZBP1 is a molecular switch regulating influenza-induced PANoptosis and perinatal lethality during development [J]. J Biol Chem, 2020, 295(24): 8325-8330. |
| [54] | Chen XY, Dai YH, Wan XX, et al. ZBP1-mediated necroptosis: mechanisms and therapeutic implications [J]. Molecules, 2022, 28(1): 52. |
| [55] | Zhan JH, Wang JS, Liang YQ, et al. Apoptosis dysfunction: unravelling the interplay between ZBP1 activation and viral invasion in innate immune responses [J]. Cell Commun Signal, 2024, 22(1): 149. |
| [56] | Karki R, Sundaram B, Sharma BR, et al. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis [J]. Cell Rep, 2021, 37(3): 109858. |
| [57] | Jiao HP, Wachsmuth L, Wolf S, et al. ADAR1 averts fatal type I interferon induction by ZBP1 [J]. Nature, 2022, 607(7920): 776-783. |
| [58] | de Reuver R, Verdonck S, Dierick E, et al. ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation [J]. Nature, 2022, 607(7920): 784-789. |
| [59] | Zhang YG, Zhang JY, Xue YC. ADAR1: a mast regulator of aging and immunity [J]. Signal Transduct Target Ther, 2023, 8(1): 7. |
| [1] | WANG Yi-fan, ZHU Hong-liang. Research Progress in the Function and Their Action Mechanism of Plant PPR Protein [J]. Biotechnology Bulletin, 2025, 41(6): 27-37. |
| [2] | LI Xin-ying, SUN Jing, LYU Ruo-tong, REN Ya-juan, LUO Lei, AI Peng-fei, WANG Yan-wei. Research Progress in the Molecular Mechanism of PPR Protein-regulated Chloroplast RNA Editing [J]. Biotechnology Bulletin, 2025, 41(10): 32-42. |
| [3] | YANG Wei, GUAN Hai-feng, REN Xin-hui, PENG Jin-ju, CHEN Zhi-bao. Alleviating Effect of Astaxanthin on Liver Injury Induced by Aflatoxin B 1 and Its Mechanism [J]. Biotechnology Bulletin, 2025, 41(10): 334-342. |
| [4] | JIN Bo-yang, QIN Shi-yu, ZHANG Ming-da, LI Qian-qian, WEN Jing, SHEN Xiu-li, DU Zhi-qiang. Research on the Molecular Mechanism of Crayfish prx 6 in the Process of Defending against Staphylococcus aureus Infection [J]. Biotechnology Bulletin, 2024, 40(7): 314-322. |
| [5] | ZHAO Hong-yuan, LIU Qiang, CHENG Wen-yu. Research Progress in cGAS-STING Signaling Pathway in ASFV Antagonizing Host [J]. Biotechnology Bulletin, 2024, 40(3): 109-117. |
| [6] | LEI Qi-yi, XU Yang, LI Peng-fei. Influence and Mechanism of Bacteroides fragilis Type VI Secretory System on the Intestinal Barrier [J]. Biotechnology Bulletin, 2024, 40(3): 286-295. |
| [7] | MA Wen-ao, YANG Wei, LI Ying-chun, ZHU Yan-bin, CHEN Zhi-bao, LIU Na. Alleviating Roles and Mechanisms of Ethanol Extract from Inonotus obliquus to Intestinal Injury Induced by Lipopolysaccharide [J]. Biotechnology Bulletin, 2024, 40(12): 299-308. |
| [8] | LONG Jia-jia, LIU Wei-wei, FAN Xin-hao, LI Wang-chang, YANG Xiao-gan, TANG Zhong-lin. Study on Constructing an RNA Editing Map of Pig Based on Large-scale RNA-seq Data [J]. Biotechnology Bulletin, 2024, 40(10): 288-295. |
| [9] | JIAO Shuai, FU Yu-ze, CUI Kai, ZHANG Ji-xian, WANG Jie, BI Yan-liang, DIAO Qi-yu, ZHANG Jian-xin, ZHANG Nai-feng. Effects of Bacillus pumilus on the Intestinal Inflammation and Barrier Function of Goat Kids [J]. Biotechnology Bulletin, 2024, 40(1): 344-352. |
| [10] | YE Hong, WANG Yu-kun. Research Progress in Immune Receptor Functions of Pattern-Recognition Receptor in Plants [J]. Biotechnology Bulletin, 2023, 39(12): 1-15. |
| [11] | CHEN Ying, WANG Yi-lei, ZOU Peng-fei. Cloning and Expression Analysis of TRAF6 from Large Yellow Croaker Larimichthys crocea [J]. Biotechnology Bulletin, 2022, 38(8): 233-243. |
| [12] | ZHU Lin, XIAN Feng-jun, ZHANG Qian-nan, HU Jun. Research Progress of RNA Editing [J]. Biotechnology Bulletin, 2022, 38(1): 1-14. |
| [13] | ZOU Chen-chen, RUAN Ling-wei, SHI Hong. Wnt Signaling Pathway and Innate Immunity of Invertebrate [J]. Biotechnology Bulletin, 2021, 37(5): 182-196. |
| [14] | LI Kai-qing, LI Ying, WANG Yi-lei, ZOU Peng-fei. The Function of Receptor-interacting Protein(RIP)Kinases and the Research Progress in Teleost Fish [J]. Biotechnology Bulletin, 2021, 37(5): 197-211. |
| [15] | CHEN Min-jie, TANG Gui-yue, HONG Xiang-na, HAO Pei, JIANG Jing, LI Xuan. Research Progress on CRISPR-Cas13-mediated RNA Editing System [J]. Biotechnology Bulletin, 2020, 36(3): 1-8. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||