








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (3): 286-295.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0840
Previous Articles Next Articles
LEI Qi-yi( ), XU Yang(
), XU Yang( ), LI Peng-fei(
), LI Peng-fei( )
)
Received:2023-08-29
Online:2024-03-26
Published:2024-04-08
Contact:
XU Yang, LI Peng-fei
E-mail:leiqiyi0701@163.com;lpf1234@suda.edu.cn;yangxu@suda.edu.cn
LEI Qi-yi, XU Yang, LI Peng-fei. Influence and Mechanism of Bacteroides fragilis Type VI Secretory System on the Intestinal Barrier[J]. Biotechnology Bulletin, 2024, 40(3): 286-295.
| 引物名称Primer name | 引物序列Sequence | 引物用途Primer purpose |
|---|---|---|
| pLGB-13 vector | F: TCAAGTTGTGCGTGATGTG | pLGB-13载体引物 |
| R: GGATTCATACAAGCGGTC | pLGB-13 vector primer | |
| T6SS 5' flanking | F: CGCGGATCC CCGCATACGCCGGAAGT | T6SS基因上游同源臂扩增引物 |
| R: CAGTTTTAATCAGGGAACGAACA | 5' flanking amplification of T6SS genes | |
| T6SS 3' flanking | F: cgtgttcgttccctgattaaaactgTA CCTGAGCCGATACGACAAA | T6SS基因下游同源臂扩增引物 |
| R: tgcctgcagTGCTACGGGCATAAACCCT | 3' flanking amplification of T6SS genes | |
| T6SS mutation | F: TCCCAGCAGGGTAGCGTC | T6SS缺陷株筛选引物 |
| R: AGTGCATTCTGAATGTCCGTATT | Mutant selection | |
| Claudin1 | F: GGGGACAACATCGTGACCG | Claudin1荧光定量引物 |
| R: AGGAGTCGAAGACTTTGCACT | qPCR primer for Claudin1 amplification | |
| Claudin2 | F: CAACTGGTGGGCTACATCCTA | Claudin2荧光定量引物 |
| R: CCCTTGGAAAAGCCAACCG | qPCR primer for Claudin2 amplification | |
| Claudin3 | F: ACCAACTGCGTACAAGACGAG | Claudin3荧光定量引物 |
| R: CAGAGCCGCCAACAGGAAA | qPCR primer for Claudin3 amplification | |
| Claudin4 | F: TGGAGGACGAGACCGTCAA | Claudin4荧光定量引物 |
| R: CACGGGCACCATAATCAGCA | qPCR primer for Claudin4 amplification | |
| Occludin | F: TTGAAAGTCCACCTCCTTACAGA | Occludin荧光定量引物 |
| R: CCGGATAAAAAGAGTACGCTGG | qPCR primer for Occludin amplification | |
| ZO-1 | F: GCTTTAGCGAACAGAAGGAGC | ZO-1荧光定量引物 |
| R: TTCATTTTTCCGAGACTTCACCA | qPCR primer for ZO-1 amplification | |
| Jam | F: TCACGTTCAGTTCTGTGACCC | Jam荧光定量引物 |
| R: TGACCTCCCCGTAGTTCTGG | qPCR primer for Jam amplification | |
| Muc1 | F: GGCATTCGGGCTCCTTTCTT | Muc1荧光定量引物 |
| R: TGGAGTGGTAGTCGATGCTAAG | qPCR primer for Muc1 amplification | |
| Gapdh | F: GTCTCCTCTGACTTCAACAGCG | Gapdh荧光定量引物 |
| R: ACCACCCTGTTGCTGTAGCCAA | qPCR primer for Gapdh amplification |
Table 1 Primer sequence
| 引物名称Primer name | 引物序列Sequence | 引物用途Primer purpose |
|---|---|---|
| pLGB-13 vector | F: TCAAGTTGTGCGTGATGTG | pLGB-13载体引物 |
| R: GGATTCATACAAGCGGTC | pLGB-13 vector primer | |
| T6SS 5' flanking | F: CGCGGATCC CCGCATACGCCGGAAGT | T6SS基因上游同源臂扩增引物 |
| R: CAGTTTTAATCAGGGAACGAACA | 5' flanking amplification of T6SS genes | |
| T6SS 3' flanking | F: cgtgttcgttccctgattaaaactgTA CCTGAGCCGATACGACAAA | T6SS基因下游同源臂扩增引物 |
| R: tgcctgcagTGCTACGGGCATAAACCCT | 3' flanking amplification of T6SS genes | |
| T6SS mutation | F: TCCCAGCAGGGTAGCGTC | T6SS缺陷株筛选引物 |
| R: AGTGCATTCTGAATGTCCGTATT | Mutant selection | |
| Claudin1 | F: GGGGACAACATCGTGACCG | Claudin1荧光定量引物 |
| R: AGGAGTCGAAGACTTTGCACT | qPCR primer for Claudin1 amplification | |
| Claudin2 | F: CAACTGGTGGGCTACATCCTA | Claudin2荧光定量引物 |
| R: CCCTTGGAAAAGCCAACCG | qPCR primer for Claudin2 amplification | |
| Claudin3 | F: ACCAACTGCGTACAAGACGAG | Claudin3荧光定量引物 |
| R: CAGAGCCGCCAACAGGAAA | qPCR primer for Claudin3 amplification | |
| Claudin4 | F: TGGAGGACGAGACCGTCAA | Claudin4荧光定量引物 |
| R: CACGGGCACCATAATCAGCA | qPCR primer for Claudin4 amplification | |
| Occludin | F: TTGAAAGTCCACCTCCTTACAGA | Occludin荧光定量引物 |
| R: CCGGATAAAAAGAGTACGCTGG | qPCR primer for Occludin amplification | |
| ZO-1 | F: GCTTTAGCGAACAGAAGGAGC | ZO-1荧光定量引物 |
| R: TTCATTTTTCCGAGACTTCACCA | qPCR primer for ZO-1 amplification | |
| Jam | F: TCACGTTCAGTTCTGTGACCC | Jam荧光定量引物 |
| R: TGACCTCCCCGTAGTTCTGG | qPCR primer for Jam amplification | |
| Muc1 | F: GGCATTCGGGCTCCTTTCTT | Muc1荧光定量引物 |
| R: TGGAGTGGTAGTCGATGCTAAG | qPCR primer for Muc1 amplification | |
| Gapdh | F: GTCTCCTCTGACTTCAACAGCG | Gapdh荧光定量引物 |
| R: ACCACCCTGTTGCTGTAGCCAA | qPCR primer for Gapdh amplification |
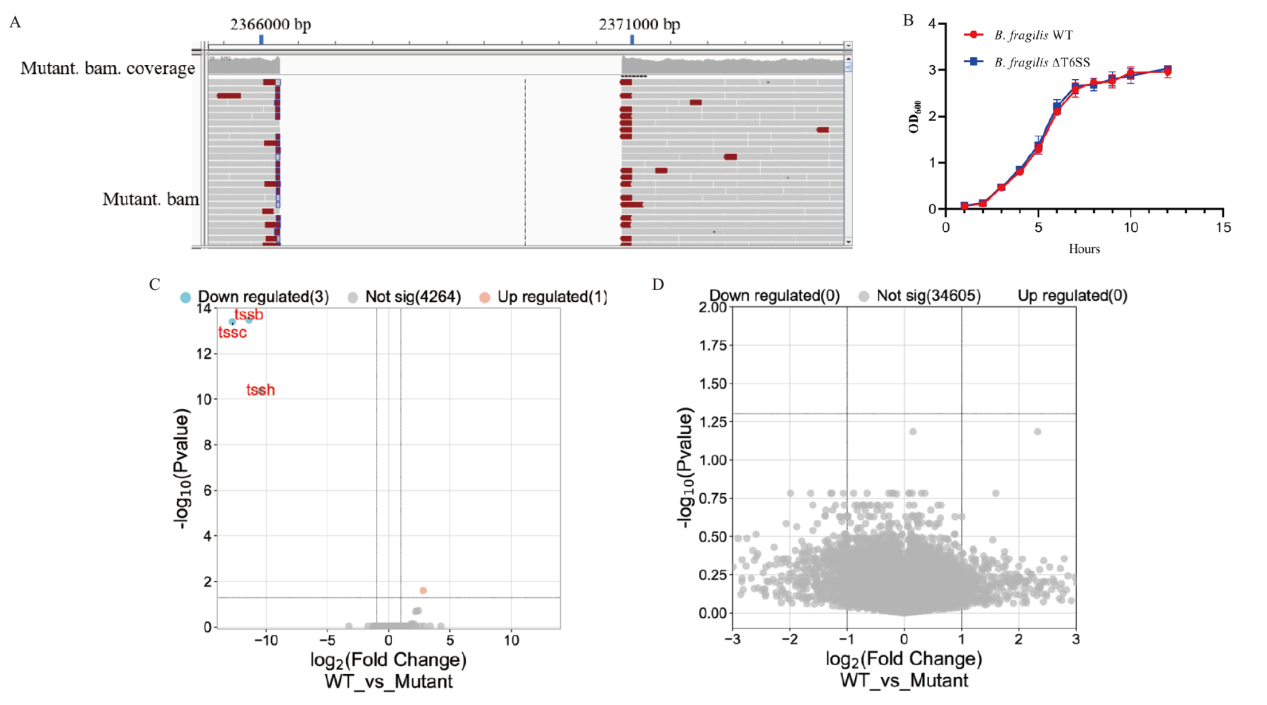
Fig. 2 Construction of B. fragilis ΔT6SS strain A: Whole genome sequencing showing deletion of tssB, tssC and tssH. B: Growth curve of WT and mutated B. fragilis strains. C-D: Bacterial transcriptomic and metabolism analysis between B. fragilis WT and T6SS mutant

Fig. 3 B. fragilis T6SS involved in protection on DSS-induced colitis A: Body weight loss; B: representative pictures of colonic tissue length; C: colon length. * P < 0. 05
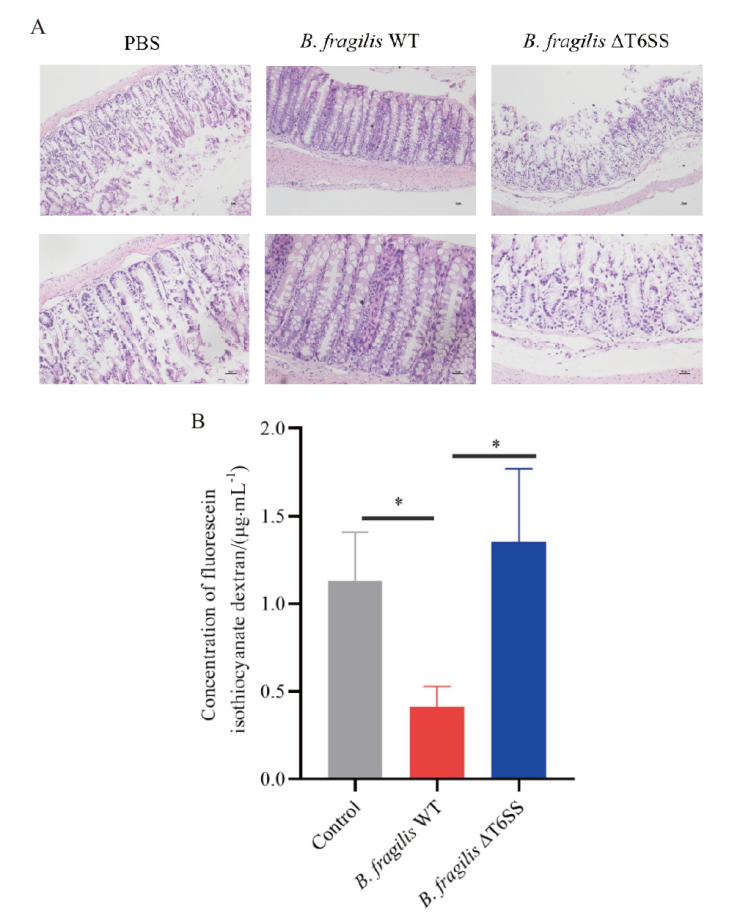
Fig. 4 B. fragilis T6SS contributes to the maintenance of the intestinal barrier in the DSS-induced colitis A: HE staining of colonic tissues in mice. B: Concentration of fluorescein isothiocyanate-dextran in mouse serum. * P < 0. 05
| [1] |
Lee SH. Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases[J]. Intest Res, 2015, 13(1): 11-18.
doi: 10.5217/ir.2015.13.1.11 pmid: 25691839 |
| [2] |
Haussner F, Chakraborty S, Halbgebauer R, et al. Challenge to the intestinal mucosa during sepsis[J]. Front Immunol, 2019, 10: 891.
doi: 10.3389/fimmu.2019.00891 pmid: 31114571 |
| [3] |
Wang YF, Li JW, Wang DP, et al. Anti-hyperglycemic agents in the adjuvant treatment of sepsis: improving intestinal barrier function[J]. Drug Des Devel Ther, 2022, 16: 1697-1711.
doi: 10.2147/DDDT.S360348 URL |
| [4] |
Naymagon S, Naymagon L, Wong SY, et al. Acute graft-versus-host disease of the gut: considerations for the gastroenterologist[J]. Nat Rev Gastroenterol Hepatol, 2017, 14(12): 711-726.
doi: 10.1038/nrgastro.2017.126 pmid: 28951581 |
| [5] |
Mathewson ND, Jenq R, Mathew AV, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease[J]. Nat Immunol, 2016, 17(5): 505-513.
doi: 10.1038/ni.3400 pmid: 26998764 |
| [6] |
Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota[J]. FEMS Microbiol Rev, 2014, 38(5): 996-1047.
doi: 10.1111/1574-6976.12075 pmid: 24861948 |
| [7] |
Kim S, Covington A, Pamer EG. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens[J]. Immunol Rev, 2017, 279(1): 90-105.
doi: 10.1111/imr.12563 pmid: 28856737 |
| [8] | Zafar H, Jr Saier MH. Gut Bacteroides species in health and disease[J]. Gut Microbes, 2021, 13(1): 1-20. |
| [9] |
Coyne MJ, Roelofs KG, Comstock LE. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements[J]. BMC Genomics, 2016, 17: 58.
doi: 10.1186/s12864-016-2377-z pmid: 26768901 |
| [10] |
Wang J, Brodmann M, Basler M. Assembly and subcellular localization of bacterial type VI secretion systems[J]. Annu Rev Microbiol, 2019, 73: 621-638.
doi: 10.1146/annurev-micro-020518-115420 pmid: 31226022 |
| [11] |
García-Bayona L, Comstock LE. Bacterial antagonism in host-associated microbial communities[J]. Science, 2018, 361(6408): eaat2456.
doi: 10.1126/science.aat2456 URL |
| [12] |
Kostow N, Welch MD. Plasma membrane protrusions mediate host cell-cell fusion induced by Burkholderia thailandensis[J]. Mol Biol Cell, 2022, 33(8): ar70.
doi: 10.1091/mbc.E22-02-0056 URL |
| [13] |
Qin L, Wang XQ, Gao YL, et al. Roles of EvpP in Edwardsiella piscicida- macrophage interactions[J]. Front Cell Infect Microbiol, 2020, 10: 53.
doi: 10.3389/fcimb.2020.00053 URL |
| [14] | Logan SL, Thomas J, Yan JY, et al. The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts[J]. Proc Natl Acad Sci USA, 2018, 115(16): E3779-E3787. |
| [15] | Coyne MJ, Zitomersky NL, McGuire AM, et al. Evidence of extensive DNA transfer between bacteroidales species within the human gut[J]. mBio, 2014, 5(3): e01305-e01314. |
| [16] | García-Bayona L, Comstock LE. Streamlined genetic manipulation of diverse Bacteroides and Parabacteroides isolates from the human gut microbiota[J]. mBio, 2019, 10(4): e01762-19. |
| [17] | Ito T, Gallegos R, Matano LM, et al. Genetic and biochemical analysis of anaerobic respiration in Bacteroides fragilis and its importance in vivo[J]. mBio, 2020, 11(1): e03238-19. |
| [18] |
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11[J]. Mol Biol Evol, 2021, 38(7): 3022-3027.
doi: 10.1093/molbev/msab120 pmid: 33892491 |
| [19] |
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees[J]. Mol Biol Evol, 1987, 4(4): 406-425.
doi: 10.1093/oxfordjournals.molbev.a040454 pmid: 3447015 |
| [20] |
Want EJ, Wilson ID, Gika H, et al. Global metabolic profiling procedures for urine using UPLC-MS[J]. Nat Protoc, 2010, 5(6): 1005-1018.
doi: 10.1038/nprot.2010.50 pmid: 20448546 |
| [21] |
Dunn WB, Broadhurst D, Begley P, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry[J]. Nat Protoc, 2011, 6(7): 1060-1083.
doi: 10.1038/nprot.2011.335 pmid: 21720319 |
| [22] |
Rosenfeldt V, Benfeldt E, Valerius NH, et al. Effect of probiotics on gastrointestinal symptoms and small intestinal permeability in children with atopic dermatitis[J]. J Pediatr, 2004, 145(5): 612-616.
doi: 10.1016/j.jpeds.2004.06.068 URL |
| [23] |
Haroun E, Kumar PA, Saba L, et al. Intestinal barrier functions in hematologic and oncologic diseases[J]. J Transl Med, 2023, 21(1): 233.
doi: 10.1186/s12967-023-04091-w pmid: 37004099 |
| [24] |
Chen Y, Yang B, Ross RP, et al. Orally administered CLA ameliorates DSS-induced colitis in mice via intestinal barrier improvement, oxidative stress reduction, and inflammatory cytokine and gut microbiota modulation[J]. J Agric Food Chem, 2019, 67(48): 13282-13298.
doi: 10.1021/acs.jafc.9b05744 URL |
| [25] |
Martín R, Chamignon C, Mhedbi-Hajri N, et al. The potential probiotic Lactobacillus rhamnosus CNCM I-3690 strain protects the intestinal barrier by stimulating both mucus production and cytoprotective response[J]. Sci Rep, 2019, 9(1): 5398.
doi: 10.1038/s41598-019-41738-5 |
| [26] |
Sofi MH, Wu YX, Ticer T, et al. A single strain of Bacteroides fragilis protects gut integrity and reduces GVHD[J]. JCI Insight, 2021, 6(3): e136841.
doi: 10.1172/jci.insight.136841 URL |
| [27] |
Jenke A, Ruf EM, Hoppe T, et al. Bifidobacterium septicaemia in an extremely low-birthweight infant under probiotic therapy[J]. Arch Dis Child Fetal Neonatal Ed, 2012, 97(3): F217-F218.
doi: 10.1136/archdischild-2011-300838 URL |
| [28] | Mater DDG, Langella P, Corthier G, et al. A probiotic Lactobacillus strain can acquire vancomycin resistance during digestive transit in mice[J]. J Mol Microbiol Biotechnol, 2008, 14(1-3): 123-127. |
| [29] |
Wegh CAM, Geerlings SY, Knol J, et al. Postbiotics and their potential applications in early life nutrition and beyond[J]. Int J Mol Sci, 2019, 20(19): 4673.
doi: 10.3390/ijms20194673 URL |
| [30] |
Russell AB, Wexler AG, Harding BN, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism[J]. Cell Host Microbe, 2014, 16(2): 227-236.
doi: S1931-3128(14)00261-3 pmid: 25070807 |
| [31] |
Le NH, Pinedo V, Lopez J, et al. Killing of Gram-negative and Gram-positive bacteria by a bifunctional cell wall-targeting T6SS effector[J]. Proc Natl Acad Sci USA, 2021, 118(40): e2106555118.
doi: 10.1073/pnas.2106555118 URL |
| [32] |
Wexler AG, Bao YQ, Whitney JC, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut[J]. Proc Natl Acad Sci USA, 2016, 113(13): 3639-3644.
doi: 10.1073/pnas.1525637113 pmid: 26957597 |
| [33] |
Ting SY, Martínez-García E, Huang S, et al. Targeted depletion of bacteria from mixed populations by programmable adhesion with antagonistic competitor cells[J]. Cell Host Microbe, 2020, 28(2): 313-321.e6.
doi: 10.1016/j.chom.2020.05.006 URL |
| [1] | XU Pei-dong, YI Jian-feng, CHEN Di, PAN Lei, XIE Bing-yan, ZHAO Wen-jun. Research Progress in the Biocontrol Secondary Metabolites of Bacillus velezensis [J]. Biotechnology Bulletin, 2024, 40(3): 75-88. |
| [2] | LIU Yan, SUN Jing, GE Liang-peng, MA Ji-deng, ZHANG Jin-wei. Effects of Intestinal Microbiota on Host Adaptive Immunity [J]. Biotechnology Bulletin, 2024, 40(1): 113-126. |
| [3] | ZHANG Jin-wei, WU Yuan-xia, SUN Jing, LI Xiao-kai, LU Lu, LI Zhou-quan, GE Liang-peng. Effects of Commensal Microbiota on Intestinal Development, Metabolism, and Mitochondrial Function in Piglets [J]. Biotechnology Bulletin, 2024, 40(1): 332-343. |
| [4] | ZHOU Ai-ting, PENG Rui-qi, WANG Fang, WU Jian-rong, MA Huan-cheng. Analysis of Metabolic Differences of Biocontrol Strain DZY6715 at Different Growth Stages [J]. Biotechnology Bulletin, 2023, 39(9): 225-235. |
| [5] | SHA Shan-shan, DONG Shi-rong, YANG Yu-ju. Research Progress in Gut Microbiota and Metabolites Regulating Host Intestinal Immunity [J]. Biotechnology Bulletin, 2023, 39(8): 126-136. |
| [6] | XIONG Shu-qi. Towards the Understanding on the Physiological Functions of Bile Acids and Interactions with Gut Microbiota [J]. Biotechnology Bulletin, 2023, 39(4): 187-200. |
| [7] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [8] | WANG Song, JIAN Xiao-ping, PAN Wan-shu, ZHANG Yong-guang, WANG Tao, YOU Ling. Effects of Fermented Corn Xiaoqu Distiller's Grains Feed on the Intestinal Microbiota of Growing-Finishing Pigs [J]. Biotechnology Bulletin, 2022, 38(9): 248-257. |
| [9] | CHEN Tian-ci, WU Shao-lan, YANG Guo-hui, JIANG Dan-xia, JIANG Yu-ji, CHEN Bing-zhi. Effects of Ganoderma resinaceum Alcohol Extract on Sleep and Intestinal Microbiota in Mice [J]. Biotechnology Bulletin, 2022, 38(8): 225-232. |
| [10] | HE Ya-lun, ZENG Li-rong, LIU Xiong, ZHANG Ling, WANG Qiong. Effects of High-dose Tannic Acid on the Intestinal Barrier Function and Gut Microbiota in Mice [J]. Biotechnology Bulletin, 2022, 38(4): 278-287. |
| [11] | XIE Guo-zhen, TANG Yuan, NING Xiao-mei, QIU Ji-hui, TAN Zhou-jin. Effects of Dendrobium officinale Polysaccharides on the Intestinal Mucosal Structure and Microbiota in Mice Fed a High-fat Diet [J]. Biotechnology Bulletin, 2022, 38(2): 150-157. |
| [12] | YANG Yu-ping, ZHANG Xia, WANG Chong-chong, WANG Xiao-yan. Study on Urine Metabolomics in Rats of Different Ages [J]. Biotechnology Bulletin, 2022, 38(2): 166-172. |
| [13] | YANG Rui-xian, LIU Ping, WANG Zu-hua, RUAN Bao-shuo, WANG Zhi-da. Analysis of Antimicrobial Active Metabolites from Antagonistic Strains Against Fusarium solani [J]. Biotechnology Bulletin, 2022, 38(2): 57-66. |
| [14] | LIU Chuan-he, HE Han, HE Xiu-gu, LAI Qiu-qin, LIU Kai, SHAO Xue-hua, LAI Duo, KUANG Shi-zi, XIAO Wei-qiang. Unveiling the Mechanisms of Pineapple Responding to Anti-chilling by Gauze Covering in Winter via Transcriptome and Metabolome Profiling [J]. Biotechnology Bulletin, 2022, 38(11): 58-69. |
| [15] | LIU Chuan-he, HE Han, HE Xiu-gu, LIU Kai, SHAO Xue-hua, LAI Duo, KUANG Shi-zi, XIAO Wei-qiang. Analysis of Differential Metabolites and Bacterial Community Structure in Soils of a Pineapple Orchard in Different Continuous-cropping Years [J]. Biotechnology Bulletin, 2021, 37(8): 162-175. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||