








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (4): 101-113.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0928
SU Yan-zhu( ), LI Da, ZHANG Ai-ai, LIU Yong-guang, ZHANG Xiu-rong(
), LI Da, ZHANG Ai-ai, LIU Yong-guang, ZHANG Xiu-rong( ), XUE Qi-qin(
), XUE Qi-qin( )
)
Received:2025-08-27
Online:2026-02-09
Published:2026-02-09
Contact:
ZHANG Xiu-rong, XUE Qi-qin
E-mail:suyanz1526@163.com;zhangxiurong@wfust.edu.cn;xueqiqin@163.com
SU Yan-zhu, LI Da, ZHANG Ai-ai, LIU Yong-guang, ZHANG Xiu-rong, XUE Qi-qin. Identification and Expression Analysis of CAD Gene Family in Soybean(Glycine max (L.) Merr.)[J]. Biotechnology Bulletin, 2026, 42(4): 101-113.
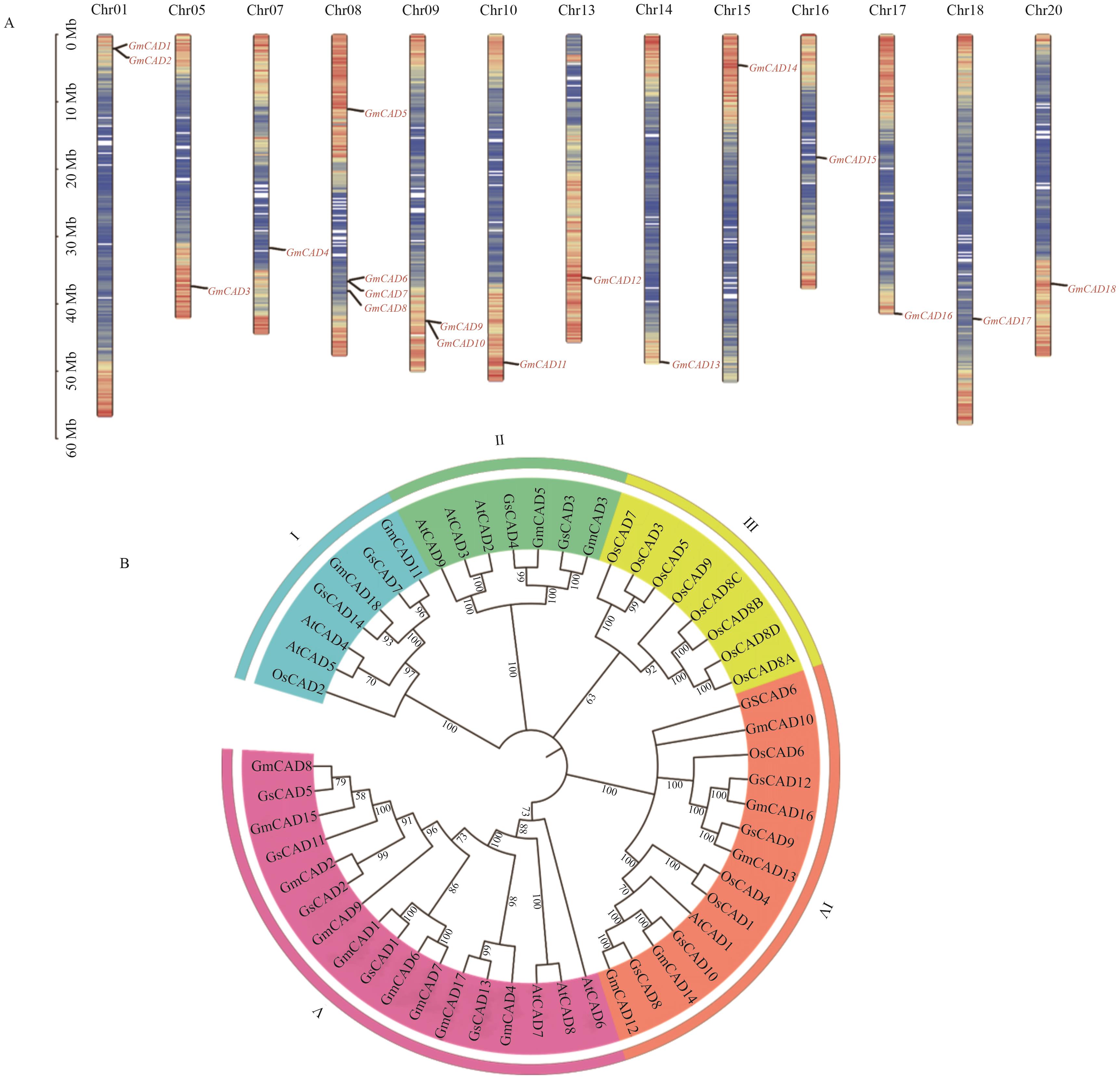
Fig. 1 Chromosomal distribution (A) and phylogenetic analysis (B) of GmCAD genesA: The colors on the chromosome indicate the density of the gene. The redder the color, the higher the density. The bluer the color, the lower the density
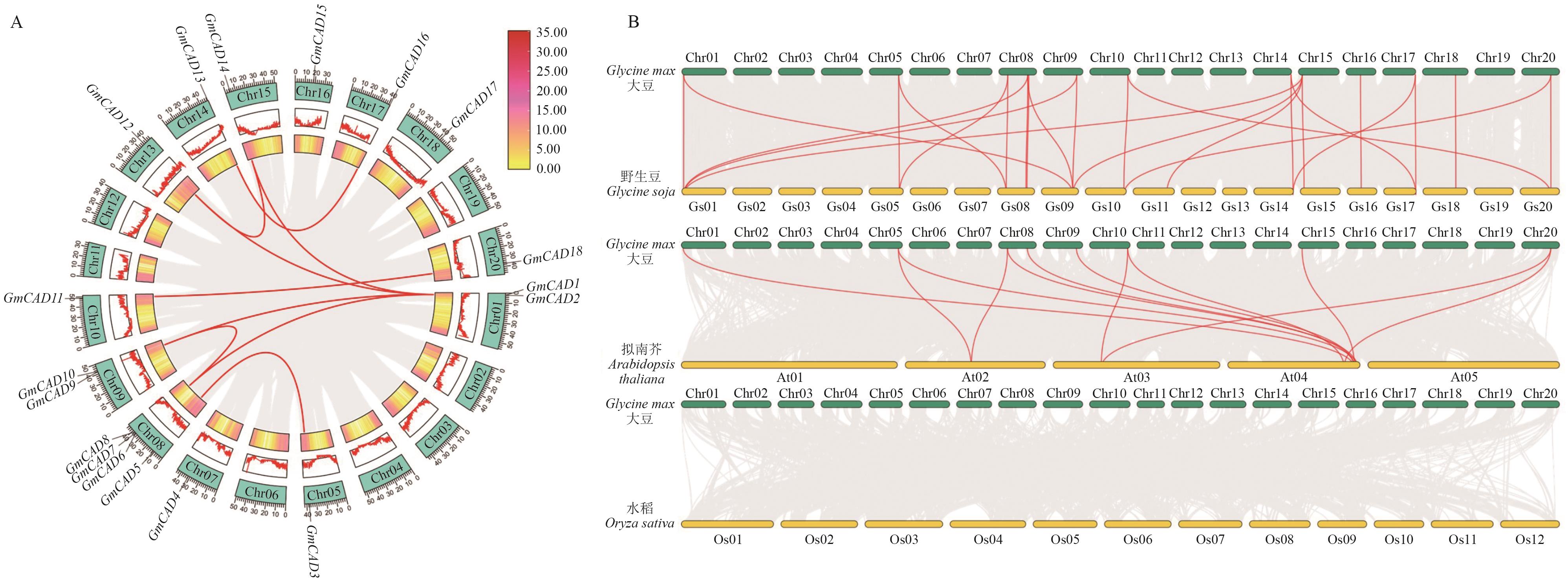
Fig. 3 Colinear analysis in the CAD gene familyA: Intraspecific colinear analysis of GmCAD genes family. Red curved line connects duplicated gene pairs. The outer part indicates the locations of these genes on the chromosome, and the green boxs correspond to different chromosomes (Chr 01-Chr 20). Red serrated lines highlight the gene density on each chromosome. The inner frames of the chart further emphasize the gene density in the chromosome, and with yellow to red indicating a change from small to large gene density. B: Colinear analysis of CAD gene between soybean and other species. The red lines highlight collinear pairs of GmCAD genes, while the gray lines in the background indicate collinear blocks between the genome of soybean and the other species
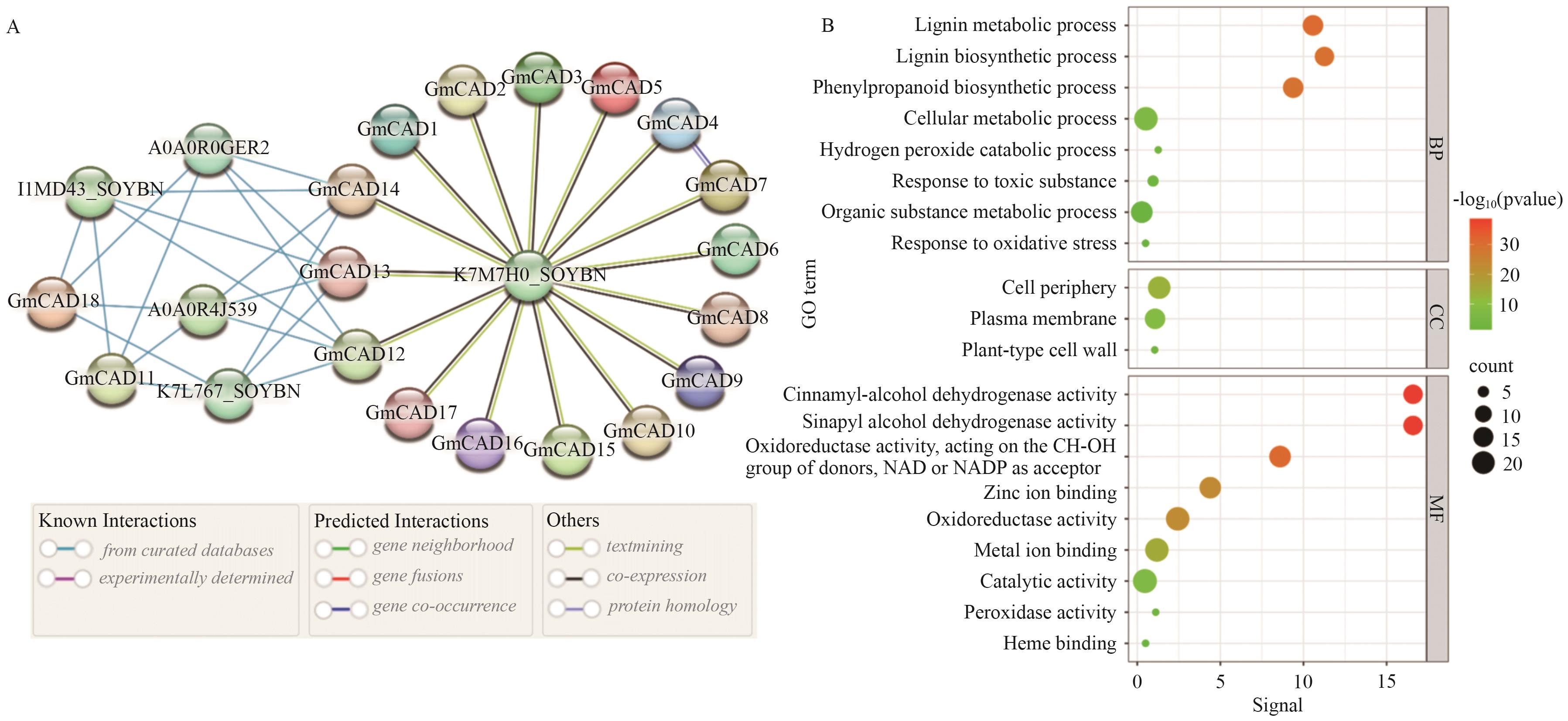
Fig. 5 Protein-protein interaction (PPI) network (A) and GO enrichment analysis (B) of GmCAD genesBP: Biological process. CC: Cellular component. MF: Molecular function
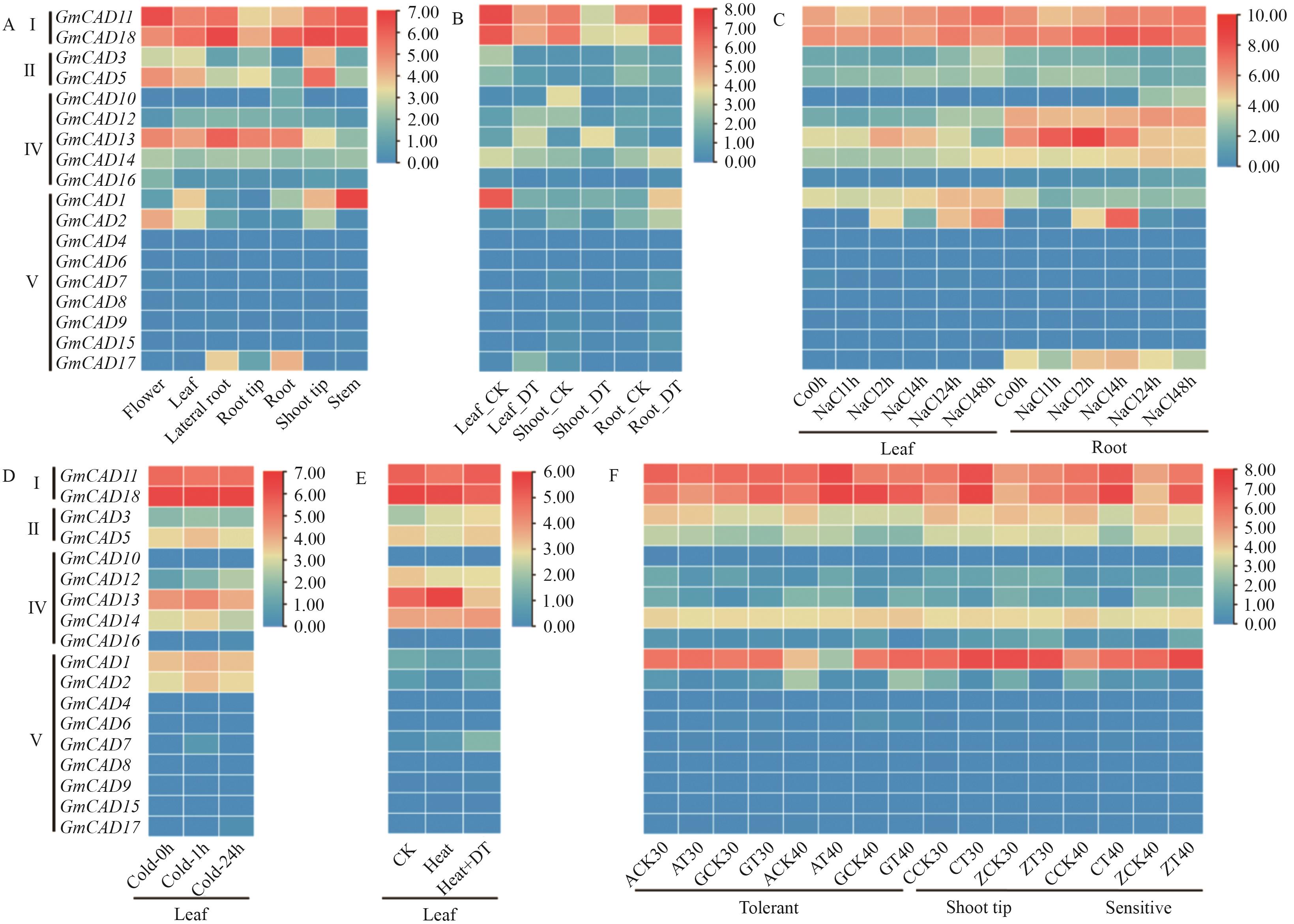
Fig. 6 Tissue-specific expression characteristics (A) and expression patterns analysis under different stress conditions (B-F) of GmCAD family genes in soybean(Glycine max (L.) Merr.)B-F: the expression patterns of the GmCAD genes under conditions of drought (DT), shade, cold, heat and salt stress, respectively. In the figures, CK refers to the control, and DT to drought stress. In Figure C, ‘A’ refers to shade-tolerant soybeans, ‘C’ to shade-intolerant soybeans, and ‘T’ to shading treatment
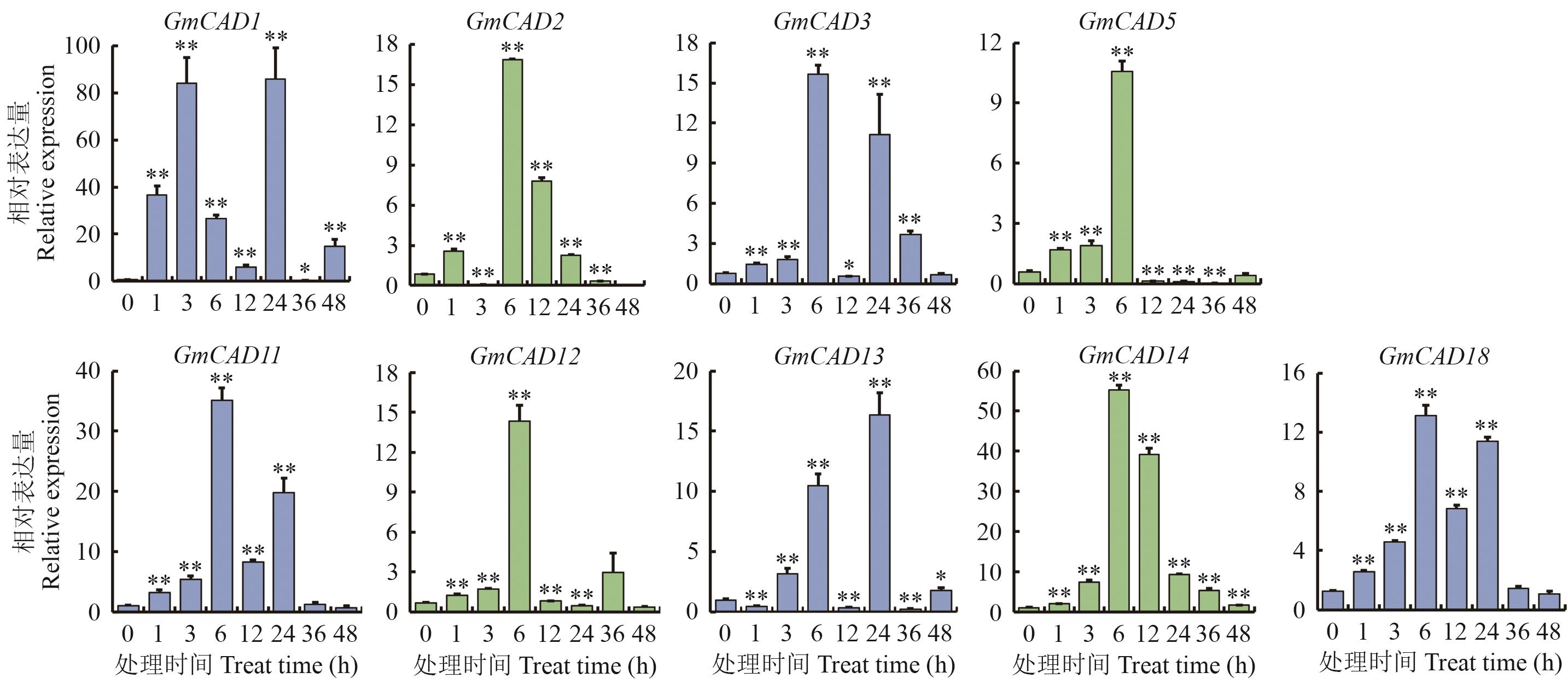
Fig. 7 The CAD genes expression under salt stress*and ** indicate that there are significant difference and extremely significant differences in expressions when comparing 1 h to 48 h after salt stress treatment with 0 before the treatment, with P<0.05 and P<0.01, the same below
| [1] | 刘欣婷, 王娟, 侯献飞, 等. 木质素及其合成基因在作物抗倒伏中的功能及其研究进展 [J]. 分子植物育种, 2019, 17(2): 655-662. |
| Liu XT, Wang J, Hou XF, et al. Progress of functions of lignin and relevant genes in plant lodging resistance [J]. Mol Plant Breed, 2019, 17(2): 655-662. | |
| [2] | 刘威. 薄皮甜瓜肉桂醇脱氢酶(CAD)在非生物胁迫下参与木质素合成的功能探究 [D]. 沈阳: 沈阳农业大学, 2019. |
| Liu W. Functional research of CmCADs in lignin biosynthesis in oriental melon (Cucumis melo L.) under abiotic stresses [D]. Shenyang: Shenyang Agricultural University, 2019. | |
| [3] | Gross GG, Stöckigt J, Mansell RL, et al. Three novel enzymes involved in the reduction of ferulic acid to coniferyl alcohol in higher plants: ferulate: Co a ligase, feruloyl-Co a reductase and coniferyl alcohol oxidoreductase [J]. FEBS Lett, 1973, 31(3): 283-286. |
| [4] | Knight ME, Halpin C, Schuch W. Identification and characterisation of cDNA clones encoding cinnamyl alcohol dehydrogenase from tobacco [J]. Plant Mol Biol, 1992, 19(5): 793-801. |
| [5] | Kim SJ, Kim MR, Bedgar DL, et al. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis [J]. Proc Natl Acad Sci U S A, 2004, 101(6): 1455-1460. |
| [6] | Tobias CM, Chow EK. Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification [J]. Planta, 2005, 220(5): 678-688. |
| [7] | Saballos A, Ejeta G, Sanchez E, et al. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [sorghum bicolor (L.) moench] identifies SbCAD2 as the brown midrib6 gene [J]. Genetics, 2009, 181(2): 783-795. |
| [8] | 任晓庆, 王波, 欧阳春平, 等. 谷子CAD基因家族的鉴定及分析 [J]. 山西农业科学, 2022, 50(3): 289-295. |
| Ren XQ, Wang B, Ouyang CP, et al. Identification and analysis of millet CAD gene family [J]. J Shanxi Agric Sci, 2022, 50(3): 289-295. | |
| [9] | An FF, Chen T, Zhu WL, et al. Systematic analysis of cinnamyl alcohol dehydrogenase family in cassava and validation of MeCAD13 and MeCAD28 in lignin synthesis and postharvest physiological deterioration [J]. Int J Mol Sci, 2024, 25(21): 11668. |
| [10] | Lu SS, Liu W, Yang LJ, et al. Transcriptome and VcCADs gene family analyses reveal mechanisms of blight resistance in rabbiteye blueberry [J]. Front Plant Sci, 2025, 16: 1601658. |
| [11] | Park HL, Kim TL, Bhoo SH, et al. Biochemical characterization of the rice cinnamyl alcohol dehydrogenase gene family [J]. Molecules, 2018, 23(10): 2659. |
| [12] | Fan L, Shi WJ, Hu WR, et al. Molecular and biochemical evidence for phenylpropanoid synthesis and presence of wall-linked phenolics in cotton fibers [J]. J Integr Plant Biol, 2009, 51(7): 626-637. |
| [13] | Guillaumie S, Pichon M, Martinant JP, et al. Differential expression of phenylpropanoid and related genes in brown-midrib bm1, bm2, bm3, and bm4 young near-isogenic maize plants [J]. Planta, 2007, 226(1): 235-250. |
| [14] | Rong W, Luo M, Shan T, et al. A wheat cinnamyl alcohol dehydrogenase TaCAD12 contributes to host resistance to the sharp eyespot disease [J]. Front Plant Sci, 2016, 7: 1723. |
| [15] | 刘堰珺, 马静, 王广龙, 等. 胡萝卜肉桂醇脱氢酶基因的克隆及其对非生物胁迫的响应 [J]. 西北植物学报, 2016, 36(7): 1294-1301. |
| Liu YJ, Ma J, Wang GL, et al. Cloning and expression profile analysis of the gene encoding cinnamyl alcohol dehydrogenase under abiotic stress in carrot [J]. Acta Bot Boreali Occidentalia Sin, 2016, 36(7): 1294-1301. | |
| [16] | Liu W, Jin YZ, Li MM, et al. Analysis of CmCADs and three lignifying enzymes in oriental melon (‘CaiHong7’) seedlings in response to three abiotic stresses [J]. Sci Hortic, 2018, 237: 257-268. |
| [17] | Liu W, Jiang Y, Wang CH, et al. Lignin synthesized by CmCAD2 and CmCAD3 in oriental melon (Cucumis melo L.) seedlings contributes to drought tolerance [J]. Plant Mol Biol, 2020, 103(6): 689-704. |
| [18] | Raza A, Asghar MA, Javed HH, et al. Optimum nitrogen improved stem breaking resistance of intercropped soybean by modifying the stem anatomical structure and lignin metabolism [J]. Plant Physiol Biochem, 2023, 199: 107720. |
| [19] | Gu Y, Zheng HY, Li S, et al. Effects of narrow-wide row planting patterns on canopy photosynthetic characteristics, bending resistance and yield of soybean in maize-soybean intercropping systems [J]. Sci Rep, 2024, 14: 9361. |
| [20] | 孙洪吉. 大豆GmCAD1基因的抗旱功能研究 [D]. 长春: 吉林农业大学, 2024. |
| Sun HJ. Study on the drought resistance function of soybean GmCAD1 gene [D]. Changchun: Jilin Agricultural University, 2024. | |
| [21] | 金贺. 大豆胞囊线虫胁迫下微紫青霉处理对GmC4H和GmCAD的影响及抗性机制研究 [D]. 沈阳: 沈阳农业大学, 2022. |
| Jin H. Effects of Penicillium purpureus on GmC4H and GmCAD under soybean cyst nematode stress and their resistance mechanism [D]. Shenyang: Shenyang Agricultural University, 2022. | |
| [22] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [23] | Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms [J]. Mol Biol Evol, 2018, 35(6): 1547-1549. |
| [24] | Bailey TL, Boden M, Buske FA, et al. MEME Suite: tools for motif discovery and searching [J]. Nucleic Acids Res, 2009, 37(): W202-W208. |
| [25] | Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server [J]. Bioinformatics, 2015, 31(8): 1296-1297. |
| [26] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| [27] | Almeida-Silva F, Pedrosa-Silva F, Venancio TM. The Soybean Expression Atlas v2: a comprehensive database of over 5000 RNA-seq samples [J]. Plant J, 2023, 116(4): 1041-1051. |
| [28] | Su YZ, Hao XS, Zeng WY, et al. Genome-wide association with transcriptomics reveals a shade-tolerance gene network in soybean [J]. Crop J, 2024, 12(1): 232-243. |
| [29] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [30] | 胡文冉, 郝晓燕, 赵准, 等. 棉花CAD基因家族的生物信息学分析 [J]. 生物技术进展, 2023, 13(3): 412-424. |
| Hu WR, Hao XY, Zhao Z, et al. Bioinformatic analysis of the CAD gene family in cotton [J]. Curr Biotechnol, 2023, 13(3): 412-424. | |
| [31] | Cannon SB, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana . [J]. BMC Plant Biol, 2004, 4(1): 10. |
| [32] | Sibout R, Eudes A, Mouille G, et al. CINNAMYL ALCOHOL DEHYDROGENASE-C and -d are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis [J]. Plant Cell, 2005, 17(7): 2059-2076. |
| [33] | Hirano K, Aya K, Kondo M, et al. OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm [J]. Plant Cell Rep, 2012, 31(1): 91-101. |
| [34] | 尹言言, 刘靖, 郑炳松, 等. 脱落酸调控植物非生物胁迫研究进展 [J]. 核农学报, 2025, 39(9): 1916-1927. |
| Yin YY, Liu J, Zheng BS, et al. Research progress of abscisic acid in regulating plant abiotic stress [J]. J Nucl Agric Sci, 2025, 39(9): 1916-1927. | |
| [35] | Cheng H, Li LL, Xu F, et al. Expression patterns of a cinnamyl alcohol dehydrogenase gene involved in lignin biosynthesis and environmental stress in Ginkgo biloba [J]. Mol Biol Rep, 2013, 40(1): 707-721. |
| [36] | Deng WW, Zhang M, Wu JQ, et al. Molecular cloning, functional analysis of three cinnamyl alcohol dehydrogenase (CAD) genes in the leaves of tea plant, Camellia sinensis [J]. J Plant Physiol, 2013, 170(3): 272-282. |
| [37] | Kim YH, Bae JM, Huh GH. Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweet potato in response to plant developmental stage and environmental stress [J]. Plant Cell Rep, 2010, 29(7): 779-791. |
| [38] | 龙国辉, 武鹏雨, 付嘉智, 等. 过氧化物酶调控木质素合成研究进展 [J]. 现代农业科技, 2021(23): 47-49, 54. |
| Long GH, Wu PY, Fu JZ, et al. Research progress on regulation of peroxidase on lignin synthesis [J]. Mod Agric Sci Technol, 2021(23): 47-49, 54. | |
| [39] | Barceló AR, Ros LVG, Carrasco AE. Looking for syringyl peroxidases [J]. Trends Plant Sci, 2007, 12(11): 486-491. |
| [1] | HE Qi-chen, YANG Yang, ALIYA Waili, TANG Xin-yue, LI Zhong-xi, CHEN Yong-kun, CHEN Ling-na. Investigation into the Family Characteristics of the Lavender Copper Amine Oxidase Gene and the Role of LaCuAO1 in Bioamine Degradation [J]. Biotechnology Bulletin, 2026, 42(1): 114-124. |
| [2] | YANG Dan, JIN Ya-rong, MAO Chun-li, WANG Bi-xian, ZHANG Ya-ning, YANG Zhi-yi, ZHOU Zhi-yao, YANG Rui-ming, FAN Heng-rui, HUANG Lin-kai, YAN Hai-dong. Identification and Expression Analysis of C2H2 Gene Family in Cenchrus purpureus [J]. Biotechnology Bulletin, 2026, 42(1): 251-261. |
| [3] | GAN Chen-lu, YOU Yu-ting, XIE Han-dan, ZENG Zi-xian, ZHU Bo. Research Progress in Flavin Monooxygenases in Plants [J]. Biotechnology Bulletin, 2026, 42(1): 1-12. |
| [4] | REN Yun-er, WU Guo-qiang, CHENG Bin, WEI Ming. Genome-wide Identification of the BvATGs Genes Family in Sugar Beet (Beta vulgaris L.) and Analysis of Their Expression Pattern under Salt Stress [J]. Biotechnology Bulletin, 2026, 42(1): 184-197. |
| [5] | YANG Yue-qin, XING Ying, ZHONG Zi-he, TIAN Wei-jun, YANG Xue-qing, WANG Jian-xu. Expression and Functional Analysis of OsMATE34 in Rice under Mercury Stress [J]. Biotechnology Bulletin, 2026, 42(1): 86-94. |
| [6] | LI Jian-bin, HOU Jia-e, LI Lei-lin, AI Ming-tao, LIU Tian-tai, CUI Xiu-ming, YANG Qian. Genome-wide Identification of Panax notoginseng Lipoxygenases Coupled in Response to Methyl-jasmonate and Wounding [J]. Biotechnology Bulletin, 2026, 42(1): 218-229. |
| [7] | ZHANG Yue, DAI Yue-hua, ZHANG Ying-ying, LI Ao-hui, LI Chu-hui, XUE Jin-ai, QIN Hui-bin, CHEN Yan, NIE Meng-en, ZHANG Hai-ping. Cloning and Functional Analysis of the Soybean Enoyl-CoA Reductase ECR14 Gene [J]. Biotechnology Bulletin, 2026, 42(1): 95-104. |
| [8] | CHEN Jing-huan, FANG Guo-nan, ZHU Wen-hao, YE Guang-ji, SU Wang, HE Miao-miao, YANG Sheng-long, ZHOU Yun. Starch Characterization and Related Gene Expression Analysis of Potato Germplasm Resources [J]. Biotechnology Bulletin, 2026, 42(1): 170-183. |
| [9] | LONG Lin-xi, ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun, ZOU Jian. Identification of Sunflower GH3 Gene Family and Analysis of Their Function in Flower Development [J]. Biotechnology Bulletin, 2026, 42(1): 125-138. |
| [10] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [11] | LI Ya-tao, ZHANG Zhi-peng, ZHAO Meng-yao, LYU Zhen, GAN Tian, WEI Hao, WU Shu-feng, MA Yu-chao. Whole Genome Analysis of Bradyrhizobium sp. Bd1 and the Negative Regulating Function of TetR3 during Cell Growth and Nodulation [J]. Biotechnology Bulletin, 2025, 41(9): 289-301. |
| [12] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [13] | HUANG Guo-dong, DENG Yu-xing, CHENG Hong-wei, DAN Yan-nan, ZHOU Hui-wen, WU Lan-hua. Genome-wide Identification and Expression Analysis of the ZIP Gene Family in Soybean [J]. Biotechnology Bulletin, 2025, 41(9): 71-81. |
| [14] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [15] | GUAN Zhi-hao, SHAN Zhi-yi, XIONG He, ZHAO Rui-xue. Computational Literature-based Knowledge Discovery for Soybean Coupling Traits [J]. Biotechnology Bulletin, 2025, 41(9): 345-356. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||